Sorghum, a hardy cereal crop of major importance, was domesticated in Africa and from there was spread around the world. This domesticated variety went through selection processes forming various improved cultivars. Domesticated sorghum is usually classified by race including bicolor, guinea, durra, caudatum and kafir, but can also be classified by end uses. During the domestication and improvement processes, seeds from panicles were selected for grain sorghum, sugar from stems for sweet sorghum, biomass from entire above-ground organs for forage sorghum and long and stiff primary branches from panicles for broom sorghum. Recent studies have shown that sorghum is genetically diverse (Tao et al., 2021), but surprisingly lacks signs of domestication bottleneck effect (Smith et al., 2019), despite decreased deleterious mutation load in these domesticated lines (Lozano et al., 2021). Instead a gradual decline in genetic diversity is seen (Smith et al., 2019). Further understanding of the process of sorghum domestication and selection for improvement would help breeders exploit the genetic diversity present across the sorghum lines and is of great interest to not only breeders, but also growers and scientists.
In an effort to ananlyze sorghum domestication genomically for various end uses, Wu and colleagues from Chinese Academy of the sciences, University of Chinese Academy of the Sciences, Chinese Agricultural University, and Liaoning Academy of Agricultural Sciences phenotypically and genetically assessed 445 diverse sorghum lines from around the world, which included, wild, weedy, domesticated and improved cultivars and encompassed different end uses.
The researchers confirmed significant phenotypic differences based on end uses such as large seeds for grain sorghum, long panicles for broom sorghum, and high stem water content for sweet sorghum. Evidence of frequent exchanges of genetic information between wild and cultivated lines and among subpopulations indicates that the hybridization of the end-use groups increased genetic diversity and may have efficiently eradicated deleterious mutations.
The researchers also reported a detailed characterization of DNA polymorphisms (SNPs and small indels) near Sh1 and SbTB1. Sh1 is the gene for seed shattering in sorghum (Lin et al., 2012); the recessive allele causes the mature seeds to remain attached to the panicle which is of critical importance when harvesting the sorghum. Researchers previously studied three recessive Sh1 halpotypes, all from one area in Africa, and matched them with the dominant version of the trait in wild lines, supporting the hypothesis of multiple domestication events in sorghum (Lin et al., 2012). Wu and colleagues confirmed this idea and furthered the investigation into sorghum’s domestication by investigating SbTB1 and its relationship to Sh1. SbTB1, plus a small number of other genes, helps determine the number of lateral branches in sorghum. During domestication, branch number was reduced and non-shattering was selected for cultivation. One major haplotype for SbTB1 was found in domesticated varieties of sorghum, but three haplotypes were found in wild lines. Out of the three hapoltypes in the wild accessions, one from Northeast Africa differed from the domesticated haplotype by only a single nucleotide providing strong evidence that it is the direct ancestor to the domesticated haplotype and that there is a single point of origin for SbTB1. SbTB1 and Sh1 are examples of two haplotype change models. While SbTB1 is representative of a single domestication event possibly in bicolor, Sh1 is representative of multiple domestication events and was not present in the primitive bicolor race background.
“Sorghum is the fifth most important cereal crop. During domestication and improvement, sorghum was domesticated and diversified for forage, grain, bioenergy and brooms, etc. However, knowledge of the genomic variation during the domestication and improvement of sorghum is limited. We performed a population genomics analysis of 445 sorghum accessions worldwide. Frequent genetic exchanges, genomic footprints, and candidate gene pool during sorghum domestication and improvement were identified. In addition, eight different models of haplotype changes in domestication genes were found. These results provide a molecular basis for understanding the domestication and improvement process of sorghum”. – Hao
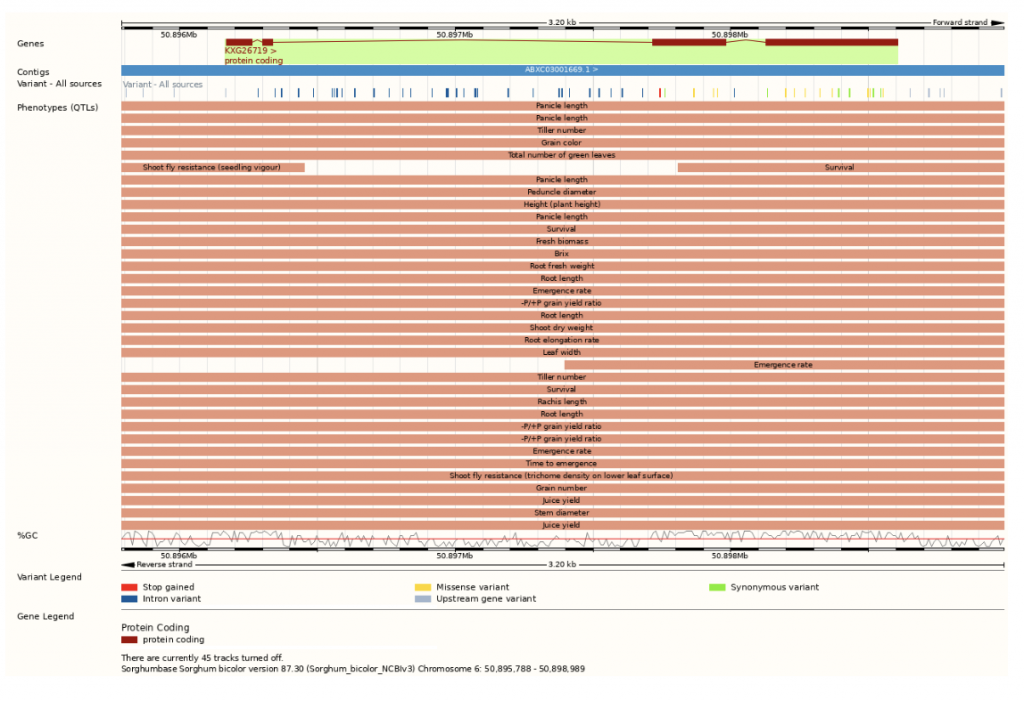
SorghumBase examples
- Sh1 – soft selection or multiple origins
- SbTB1 – hard selection or early single domestication event
- Dry – Sobic.006G147400 is a NAC gene determined to be the functional candidate of the Dry locus. It regulates stem juiciness.
SorghumBase example:
References
Lin Z, Li X, Shannon LM, Yeh CT, Wang ML, Bai G, Peng Z, Li J, Trick HN, Clemente TE, Doebley J, Schnable PS, Tuinstra MR, Tesso TT, White F, Yu J. Parallel domestication of the Shattering1 genes in cereals. Nat Genet. 2012 May 13;44(6):720-4. PMID: 2258123. DOI: 10.1038/ng.2281.
Lozano, R., Gazave, E., dos Santos, J.P.R., Stetter, M.G., Valluru, R., Bandillo, N., Fernandes, S.B., Brown, P.J., Shakoor, N., Mockler, T.C., Cooper, E.A., Perkins, M.T., Buckler, E.S., Ross-Ibarra, J., and Gore, M.A. Comparative evolutionary genetics of deleterious load in sorghum and maize. Nat. Plants 7, 17–24 (2021). PMID: 33452486. DOI: 10.1038/s41477-020-00834-5.
Smith O, Nicholson WV, Kistler L, Mace E, Clapham A, Rose P, Stevens C, Ware R, Samavedam S, Barker G, Jordan D, Fuller DQ, Allaby RG. A domestication history of dynamic adaptation and genomic deterioration in Sorghum. Nat Plants. 2019 Apr;5(4):369-379. PMID: 30962527. DOI: 10.1038/s41477-019-0397-9.
Tao, Y., Luo, H., Xu, J., Cruickshank, A., Zhao, X., Teng, F., Hathorn, A., Wu, X., Liu, Y., Shatte, T., Jordan, D., Jing, H., and Mace, E. Extensive variation within the pan-genome of cultivated and wild sorghum. Nat. Plants 7, 766–773 (2021). https://doi.org/10.1038/s41477-021-00925-x.
Wu X, Liu Y, Luo H, Shang L, Leng C, Liu Z, Li Z, Lu X, Cai H, Hao H, Jing HC. Genomic footprints of sorghum domestication and breeding selection for multiple end uses. Mol Plant. 2022 Mar 7;15(3):537-551. PMID: 34999019. DOI: 10.1016/j.molp.2022.01.002. Read more
Related Project Websites
- https://service.most.gov.cn/
- https://www.cas.cn/
- Jing lab website: http://klpr.ibcas.ac.cn/en/faculty/professor/201306/t20130604_615021.html

