S. bicolor (sorghum) and Z. mays (maize) are closely related grasses which evolved from a common ancestor approximately 12 million years ago. They differ in some significant traits such as drought and temperature tolerance, which are “mediated by specialized cell types”. Guillotin and colleagues from New York University, Cold Spring Harbor Laboratory, University of California Riverside, Michigan State University and University of Toronto compared the transcriptomes of maize and sorghum and S. viridis (millet cousin). S. viridis diverged from its common ancestor with maize and sorghum 30 million years ago; maize and sorghum then diverged from each other 5-12 million years ago. The scientists compared data from single-cell and single-nucleus in Arabidopsis thaliana or maize and discovered that single-cell provides greater depth and double the number of nuclei needed to discover the same number of clusters as with the single-cell approach. However, single-nucleus profiling provided clusters undiscovered by single-cell in tissues that were difficult to enzymatically digest. Thus, the two profiling approaches should be used in a complimentary fashion for the most accurate analysis. The researchers used both methods in this study comparing sorghum, maize and setaria, used cell-type specific marker genes from maize to predict cell identity in the other species and search for conserved gene expression. Using a stringent regulon analysis, they found 15 transcription factors and putative targets that were conserved across the three grasses. The researchers also investigated neofunctionalization of maize homeologs and identified root cell types that changed the most in maize and sorghum relative to setaria. Single cell and nuclei based pan-transcriptomes promise to reveal complex gene function and regulation as the technology is applied more broadly in crop research.
SorghumBase example:
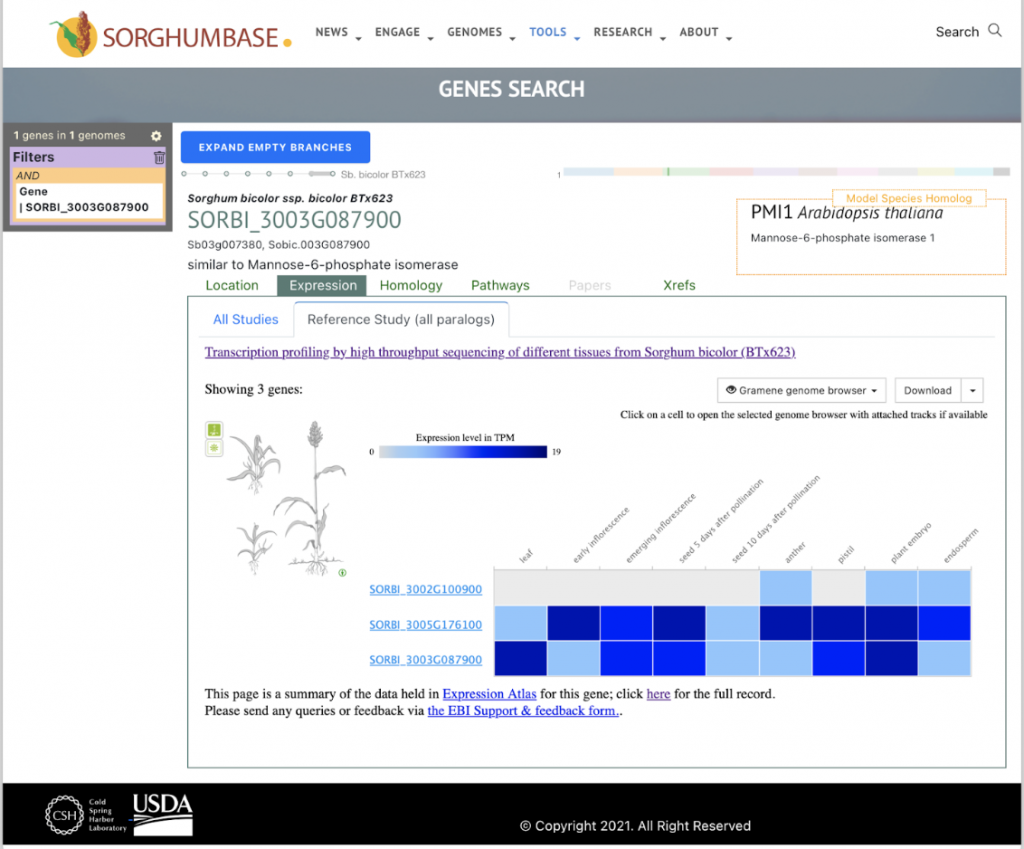
Reference:
Guillotin B, Rahni R, Passalacqua M, Mohammed MA, Xu X, Raju SK, Ramírez CO, Jackson D, Groen SC, Gillis J, Birnbaum KD. A pan-grass transcriptome reveals patterns of cellular divergence in crops. Nature. 2023 May;617(7962):785-791. PMID: 37165193. DOI: 10.1038/s41586-023-06053-0. See more
Related Project Website:
Birnbaum lab at NYU: https://pages.nyu.edu/birnbaum/