Sorghum, a crucial cereal crop with diverse applications ranging from food and feed to biofuels and alcoholic beverages, faces a significant threat from anthracnose, a devastating disease caused by the fungus Colletotrichum sublineola (Cs). Scientists from Purdue University have identified and characterized two dominant resistance genes—ANTHRACNOSE RESISTANCE GENES 4 and 5 (ARG4 and ARG5). These genes, encoding canonical nucleotide-binding leucine-rich repeat (NLR) receptors, showcase remarkable potential for broad-spectrum resistance to Cs, marking a significant leap forward in the fight against this agricultural menace. Sobic.008G166400 is the most likely ARG4 gene conferring anthracnose resistance in SAP135 and designated as ANTHRACNOSE RESISTANCE GENE 4 (ARG4) gene and Sobic.008G177900 is the ARG5 gene conferring resistance in P9830.
ARG4 and ARG5, discovered in sorghum lines SAP135 and P9830, respectively, were found to confer robust resistance to multiple strains of Cs. The study utilized sophisticated genetic mapping techniques, molecular markers, comparative genomics, and gene expression studies to pinpoint the location and function of these resistance genes. Interestingly, both ARG4 and ARG5 are nestled within clusters of duplicate NLR genes, suggesting a complex genetic landscape separated by approximately 1 Mb genomic regions. SAP135 and P9830, the resistant sorghum lines, each carry only one of the ARG genes while possessing a recessive allele at the second locus. Furthermore, the study revealed that the resistant parents and their recombinant inbred lines carrying either ARG4 or ARG5 demonstrated resistance not only to Cs strains but also exhibited overlapping specificities against different anthracnose strains.
The research validated the crucial role of ARG4 and ARG5 in anthracnose resistance through the use of sorghum lines carrying independent recessive alleles, which exhibited increased susceptibility to the fungal pathogen. These findings provide a foundation for genetic studies and resistance breeding initiatives, offering opportunities for improved crop protection strategies.
Pathogens neutralize the genetic improvements achieved through breeding. Climate change exacerbates this issue, as fungal diseases become a greater threat with virulent strains of pathogenic fungi emerging and spreading to new regions, overcoming host immunity. This situation necessitates the identification and stacking of new loci and genes to create resistant genotypes. This research showcases the potential of tapping into natural variation by utilizing whole genome resequencing and exploring allelic diversity at resistance loci. It marks a significant step towards crop improvement and deepens our understanding of resistance mechanisms in sorghum. – Mengiste
SorghumBase examples:
Sobic.008G166400 is the most likely ARG4 gene conferring anthracnose resistance in SAP135 and designated as ANTHRACNOSE RESISTANCE GENE 4 (ARG4) gene. This gene was used to explore SorghumBase below.
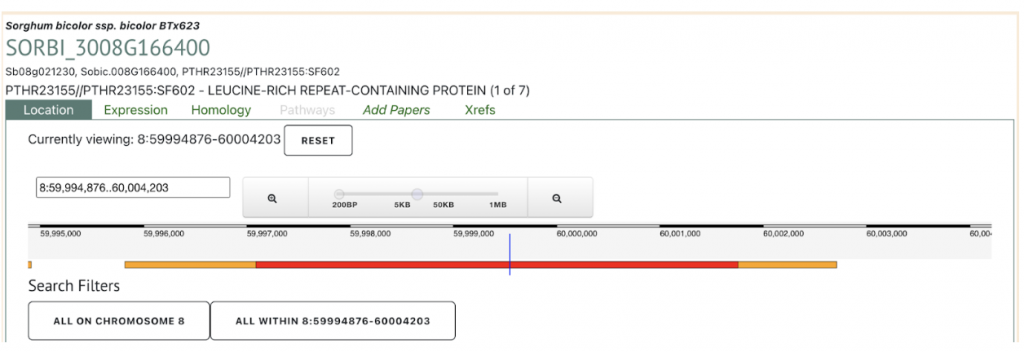
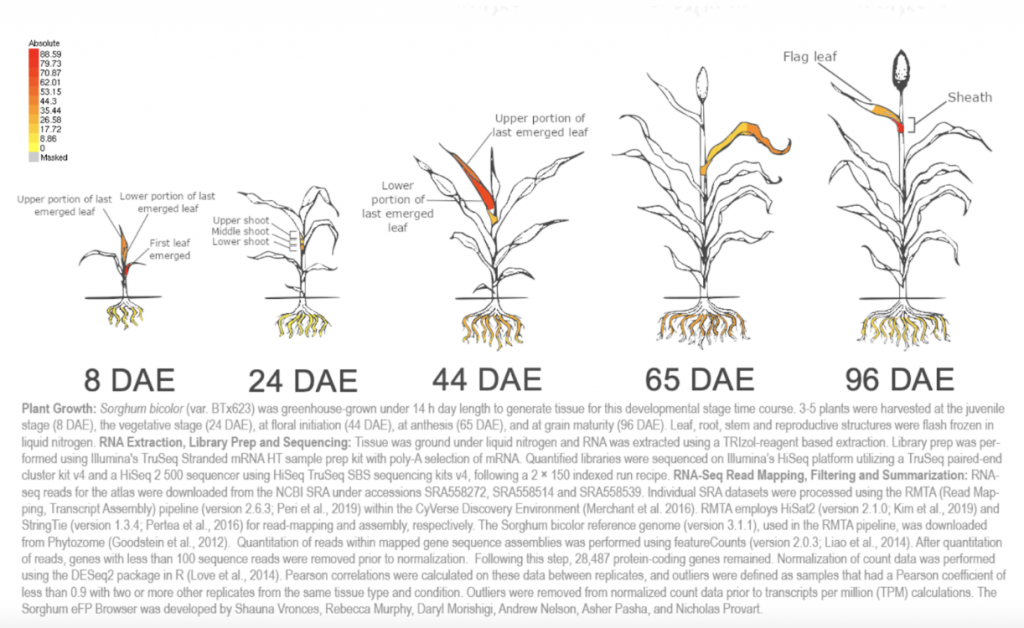


Reference:
Habte N, Girma G, Xu X, Liao CJ, Adeyanju A, Hailemariam S, Lee S, Okoye P, Ejeta G, Mengiste T. Haplotypes at the sorghum ARG4 and ARG5 NLR loci confer resistance to anthracnose. Plant J. 2023 Dec 18. PMID: 38111157. doi: 10.1111/tpj.16594. Read more
Related Project Websites:
- Mengiste Lab at Purdue University: https://ag.purdue.edu/department/btny/labs/mengiste/index.html
- Purdue Center for Global Food Security: https://www.purdue.edu/discoverypark/food/?_ga=2.142886517.1746745939.1706323565-1879135848.1706323565

