The exponential increase in sequencing data and the need for rapid processing present challenges for efficient SNP (single-nucleotide polymorphism) detection in major crops, such as sorghum, maize and soybean. SNP detection is significant for trait discovery and genomic selection, necessitating the development of faster and more efficient workflows. The researchers, from King Abdullah University of Science and Technology, University of Arizona, Huazhong Agricultural University, International Rice Research Institute, Cold Spring Harbor Laboratory, Crop Science Research Center and the USDA-ARS, introduce HPC-GVCW, an open-source high-performance computing genome variant calling workflow, designed to address these challenges. This workflow employs high-performance computing (HPC) architectures and a novel algorithm called “Genome Index splitter” to parallelize genome processing and reduce execution times significantly. By dividing genomes into smaller chunks, HPC-GVCW achieves a remarkable speedup, enabling the rapid calling of SNPs from large datasets, such as resequenced rice accessions, in a fraction of the time previously required.
The paper demonstrates the utility of HPC-GVCW by running it on a dataset comprising 25 crop genomes and calling millions of SNPs for rice, sorghum, maize and soybean. Analysis of these SNPs reveals a substantial number of novel variations, particularly in a 16-genome rice reference panel, which could significantly contribute to molecular-assisted selection breeding programs and functional genomics studies. The authors emphasize the reproducibility of their results through containerization and highlight the scalability of HPC-GVCW across various computational platforms. They also discuss future directions, including integrating machine learning-based tools like “DeepVariant” into the workflow and expanding pan-genome strategies to other species, with the goal of enhancing genetic studies and breeding programs in major crops. Overall, HPC-GVCW represents a significant advancement in SNP detection, offering a faster and more scalable solution for exploring genetic diversity in crop plants.
Our High-performance Computing (HPC) workflow was specifically crafted to expedite variant identification in key crop species, enabling scientists to dedicate their efforts primarily to downstream analysis utilizing pan-genomic variant data. The vast potential and ample opportunities within this dataset warrant thorough exploration. While the logical structure and demonstration of the workflow was straightforward, its implementation on the HPC platform presented numerous obstacles. By collaborating closely with a team of computer scientists, we successfully navigated these challenges. Such interdisciplinary collaborations will become increasingly vital in the future. – Zhou
We are excited that our hpc workflow can be wildly adapted to rapidly call SNPs on multiple crops species on genomes that range over 10 fold in size (e.g. rice GS = 400 Mb, sorghum GS = 700 Mb, and maize GS = 2400 Mb). – Wing
SorghumBase examples:
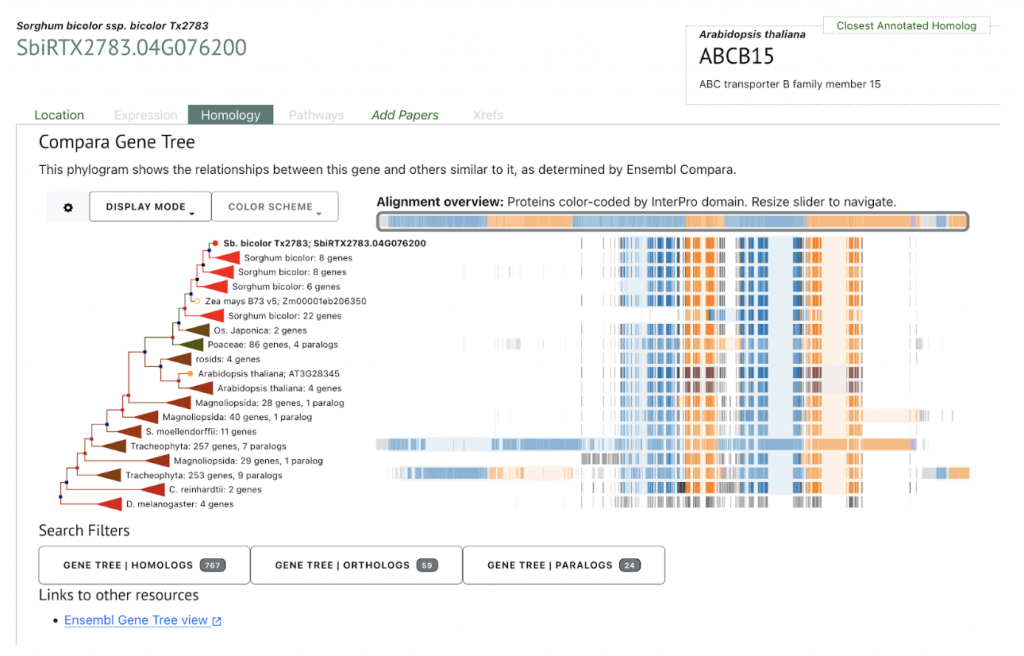
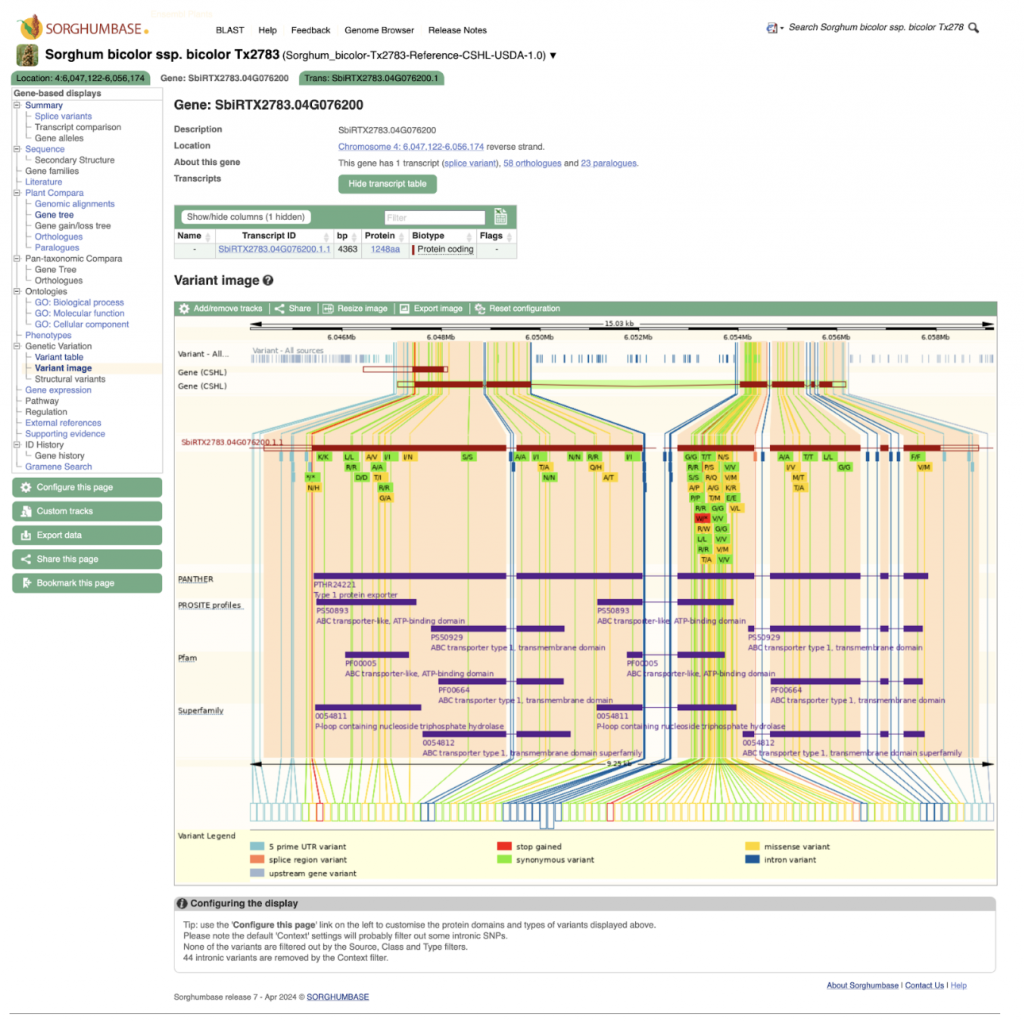
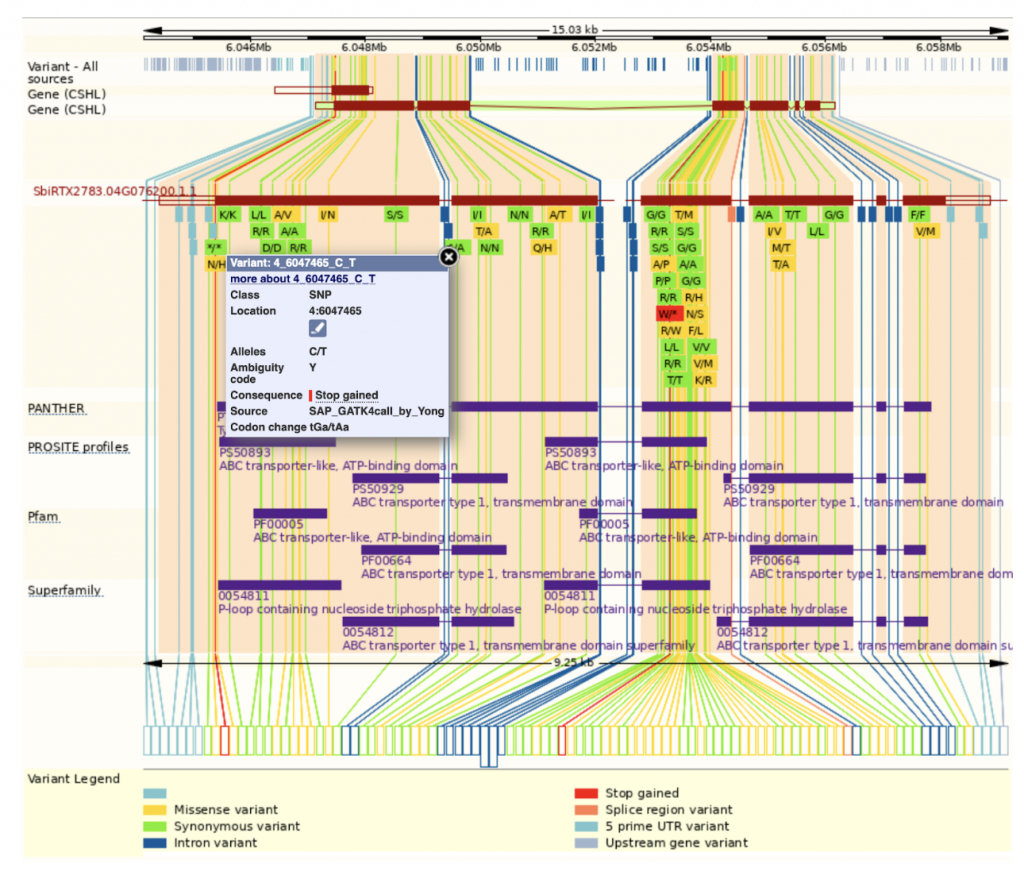
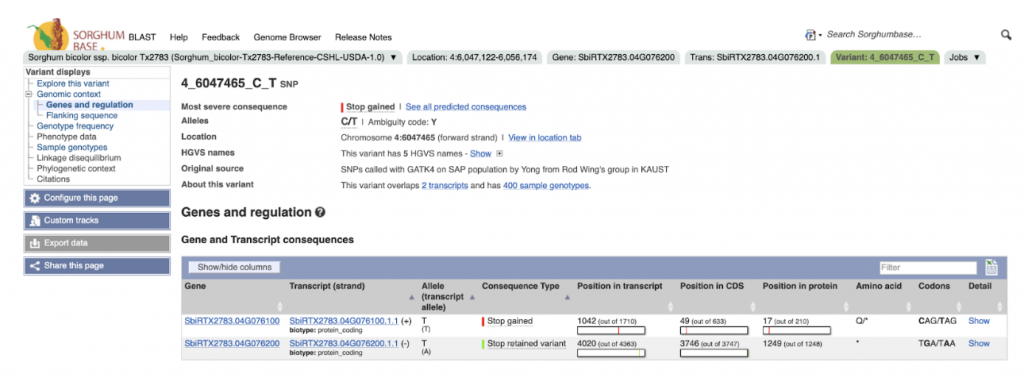
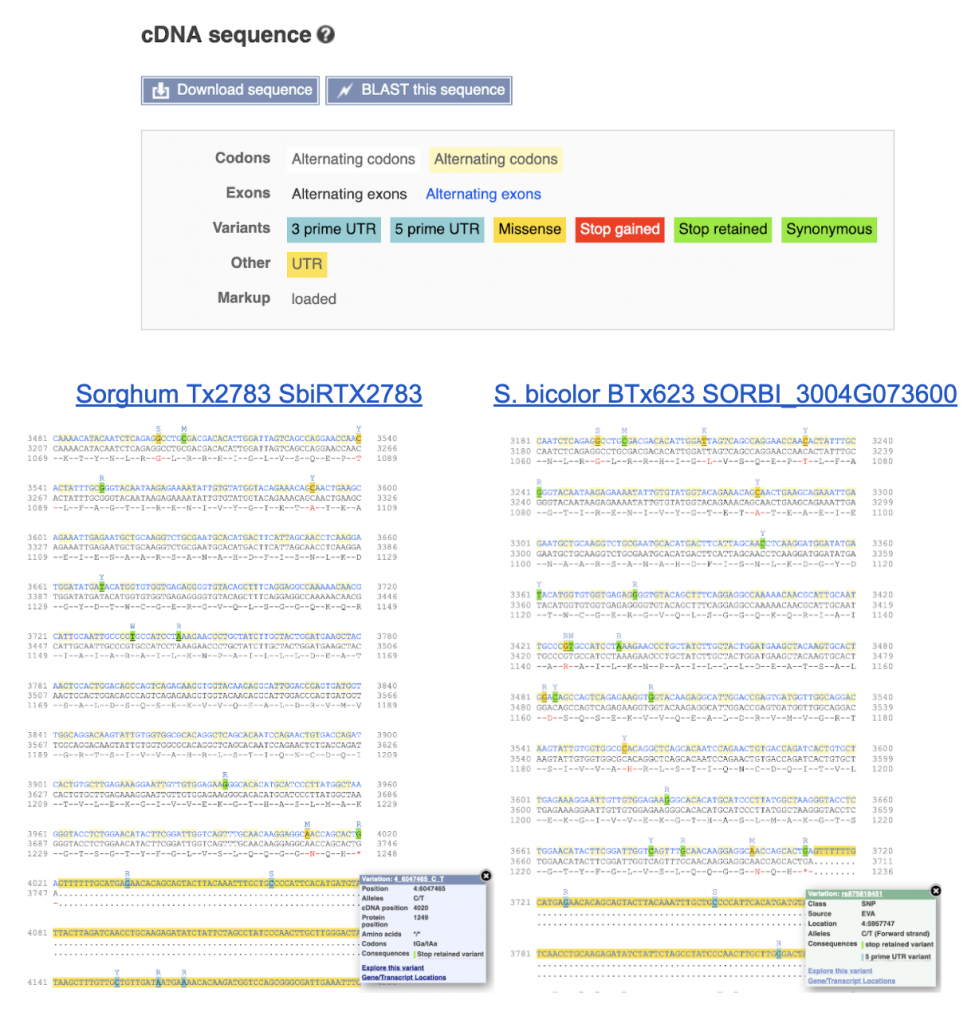
An example gene SbiRTX2783.04G076200 in the TX2783 genome is highly conserved across sorghum varieties and other crops including rice and maize, as shown in Homology view Figure 1. Homology view from the search result page on SorghumBase provides quick information on the function of this gene as ABC transporter based on homology to an arabidopsis ABC transporter with 685 identity and showing 24 paralogs in Tx2783
Reference:
Zhou Y, Kathiresan N, Yu Z, Rivera LF, Yang Y, Thimma M, Manickam K, Chebotarov D, Mauleon R, Chougule K, Wei S, Gao T, Green CD, Zuccolo A, Xie W, Ware D, Zhang J, McNally KL, Wing RA. A high-performance computational workflow to accelerate GATK SNP detection across a 25-genome dataset. BMC Biol. 2024 Jan 25;22(1):13. PMID: 38273258. doi: 10.1186/s12915-024-01820-5. Read more
Related Project Websites:
- Wing lab at the Center for Desert Agriculture at King Abdullah University of Science and Technology: https://cda.kaust.edu.sa/research/research-groups/wing-lab
- All SNP data produced for this 25-genome reference set have been publicly released through the SNP-Seek (https://snp-seek.irri.org/_download.zul), Gramene (http://ftp.gramene.org/collaborators/Yong_et_al_variation_dumps/), and KAUST Research Repository (KRR [58]) public databases for immediate access.
- Realignment data sets of near variant regions (cram file format) of the O. sativa 16-genome RPRP data set are available through Amazon Web Services (AWS) 3kricegenome bucket at SNP-Seek (https://snp-seek.irri.org/_download.zul).
- GitHub page: https://github.com/IBEXCluster/HPC-GVCW @Nagarajan Kathiresan
