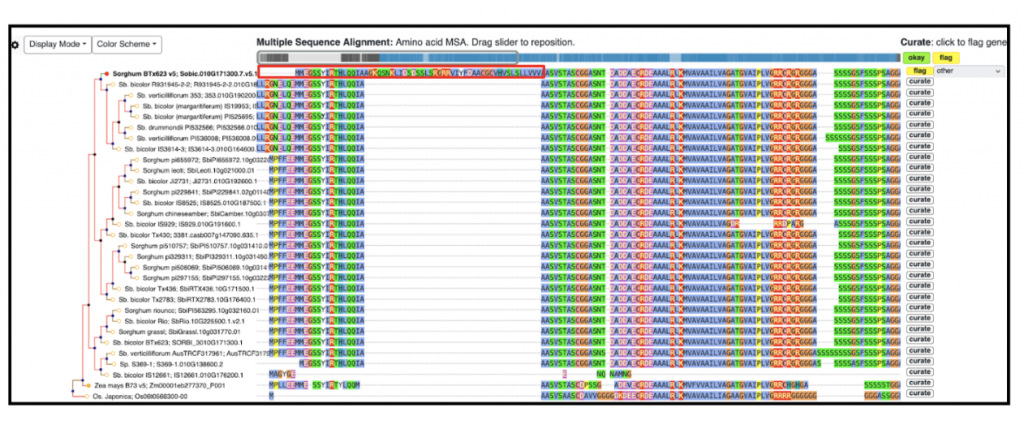
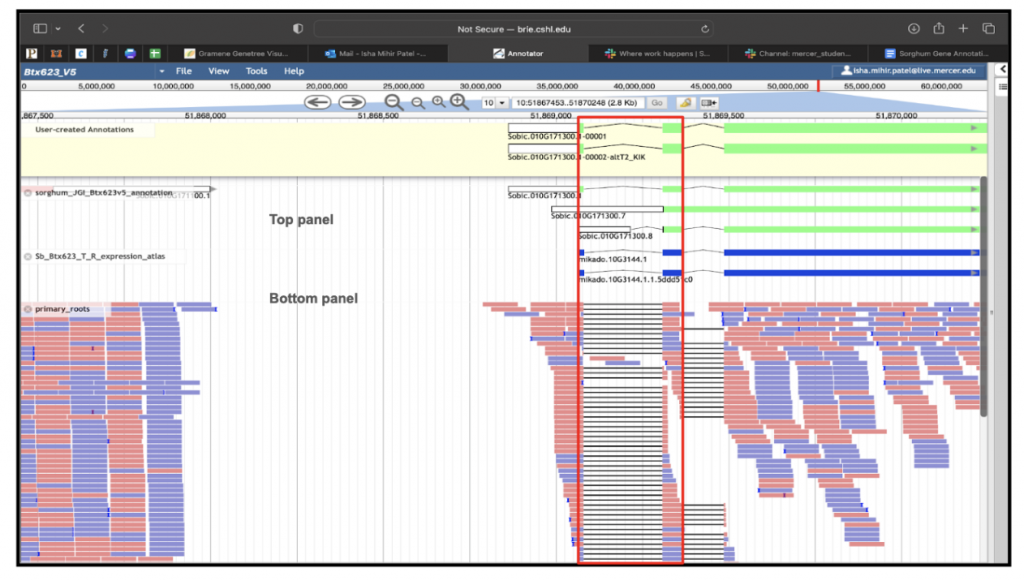
Ensuring the accuracy of gene models is crucial for designing precise experiments like CRISPR targeting. Traditionally, this validation process involves manual curation, which is time-consuming and labor-intensive as experts evaluate computational predictions against experimental evidence. However, tapping into undergraduate science majors as gene curators presents a promising solution, particularly when integrated into coursework activities. Gramene’s crowdsourcing curation initiatives have been highly valued and readily embraced, making them easily adaptable. Our ongoing collaboration with students from Mercer University focuses on structurally curating sorghum genes and annotating potentially flawed gene models using the Apollo gene editor. We developed training modules for students to curate computer-generated gene annotations over the span of 3 months. The initial module focused on training students on the Gramene Tree Tool, to curate functionally important sorghum BTx623 v5 genes. In total, the students will curate approximately 600 sorghum genes primarily identified based on their function (i.e., sugar and metal transporter genes and disease resistance genes), and based on their structure (namely split genes). We had 7 students working on the gene list that flagged 170 genes for curation. These genes were flagged based on whether the gene had gain or loss in the 5 prime, 3 prime or CDS regions. An example of gene curation (SORBI_3010G171300) is shown in Figure 1. In the second module, we developed a tutorial to guide students through the process of manual editing using the range of functionalities in the Apollo gene editor. In the module, students manually edited 31 SorghumBase genes using evidence tracks to support their edited models. See example Figure 2, Apollo curation for the flagged gene SORBI_3010G171300. Going forward the students presented their curation work at the Mercer University annual student symposium, BEAR Day (Breakthroughs in Engagement, Arts, and Research) April 18-19. Additionally, the students will work on identifying and curating sorghum orthologs of Arabidopsis genes involved in synthesis of, and response to, strigolactones. This work will be targeted towards micropublication.