Drought is a significant abiotic stress that affects crop yield and quality globally. As climate change intensifies, drought occurrences are becoming more frequent and severe. Drought stress impacts plants by disrupting physiological processes such as photosynthesis, respiration, and water retention, which in turn lead to reduced growth and yield. The molecular response of plants to drought is complex, involving numerous genes and pathways that regulate biochemical and physiological processes. For instance, water deficiency reduces photosynthesis by impairing leaf expansion and the activity of enzymes involved in the Calvin cycle. In addition to protein-coding genes, non-coding RNAs (ncRNAs), including long non-coding RNAs (lncRNAs), have been shown to play critical roles in regulating plant responses to drought and other environmental stresses.
LncRNAs, which are over 200 base pairs in length, do not code for proteins. High-throughput sequencing has identified thousands of lncRNAs in various crops, including sorghum, which is known for its drought tolerance. In this study, researchers from Shanxi Agricultural University and Heilongjiang Academy of Agricultural Sciences identified over 11,000 lncRNAs in sorghum leaves under drought conditions. Several of these lncRNAs were found to modulate stress-related genes, influencing processes like redox activity, signal transduction, and photosynthetic carbon metabolism. This research expands our understanding of lncRNAs’ roles in drought response, providing insights into how plants adapt to water scarcity at a molecular level.
SorghumBase examples:
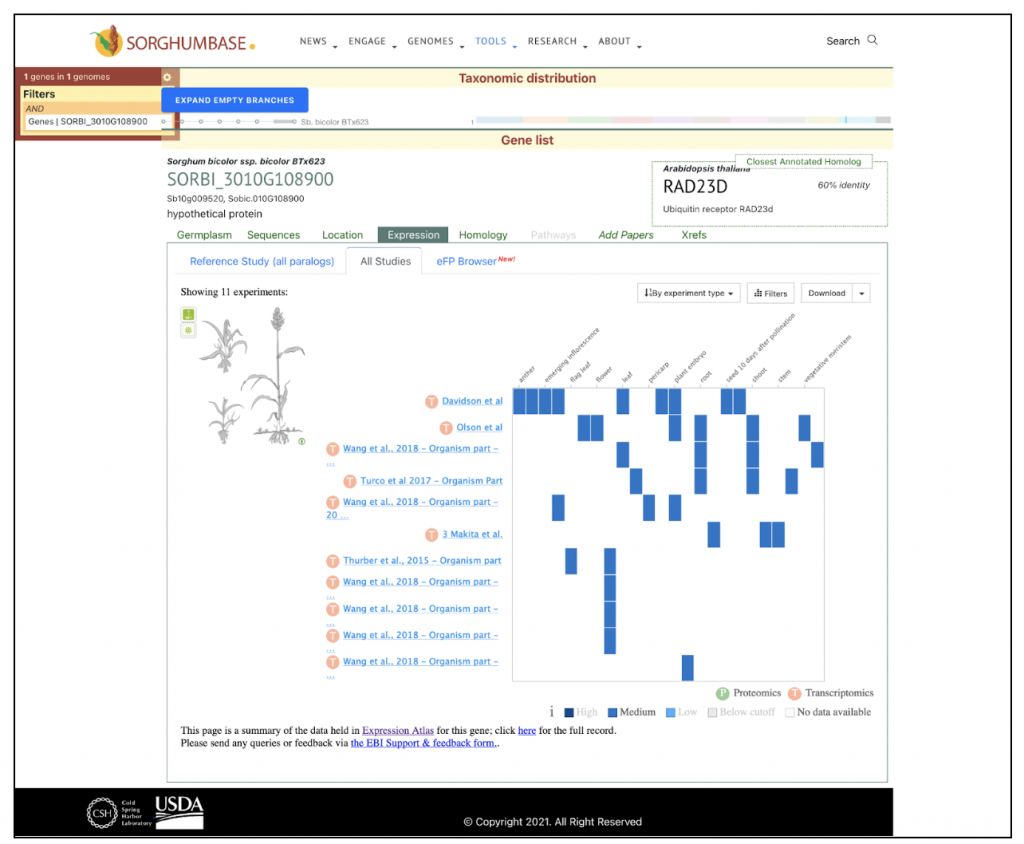
An example gene mentioned in the study, SORBI_3010G108900, is annotated as a ubiquitin receptor RAD23d-like gene. Ubiquitin receptors are integral to the ubiquitin-proteasome system (UPS), which manages the degradation of proteins that are misfolded, damaged, or no longer required. The RAD23 protein family functions by recognizing and delivering ubiquitinated proteins to the proteasome for degradation.
In this study, the expression of SORBI_3010G108900 (the ubiquitin receptor RAD23d-like gene) was found to be regulated by lncRNA TCONS_00017127. Under drought treatment DR1, this gene was downregulated more than eight-fold, suggesting that the modulation of protein degradation via the UPS, specifically through the RAD23d-like protein, plays a key role in sorghum’s drought response. The observed downregulation could indicate a reduction in the rate of protein degradation, potentially as a resource-conservation mechanism during drought stress.
The study also highlights the role of lncRNAs like TCONS_00017127 in regulating gene expression in response to drought. This finding points to a complex regulatory network that sorghum uses to adapt to environmental stresses, including drought.
Reference:
Zou C, Zhao S, Yang B, Chai W, Zhu L, Zhang C, Gai Z. Genome-wide characterization of drought-responsive long non-coding RNAs in sorghum (Sorghum bicolor). Plant Physiol Biochem. 2024 Sep;214:108908. PMID: 38976942. doi: 10.1016/j.plaphy.2024.108908. Read more
