Pearl millet is a highly resilient annual crop that thrives in diverse ecological conditions due to its extensive root system and high photosynthetic efficiency. This C4 plant contributes significantly to global terrestrial carbon sequestration and offers substantial productivity in low-nutrient soils, reducing reliance on chemical fertilizers. However, despite its exceptional nutritional value, gluten-free properties, and beneficial agronomic traits, pearl millet has not achieved widespread global popularity compared to other cereal crops. This limited popularity is primarily due to its flour’s short shelf life and the rapid development of off-flavors. The quick onset of rancidity and undesirable odors in pearl millet flour is caused by the accumulation of lipids and the enzymatic activities of phospholipases, lipoxygenases, peroxidases, and polyphenol oxidases. Additionally, external factors such as oxygen, light exposure, and storage temperatures further influence the flour’s shelf life. Oxidative rancidity, which leads to the production of hydroperoxides and secondary metabolites, contributes to the bitter and musty odors associated with pearl millet flour. To extend the shelf life of pearl millet flour, it is crucial to first assess the impact of phospholipases on hydrolytic rancidity and then manage factors contributing to oxidative rancidity. To that end, scientists from ICRISAT conducted a study which focused on a thorough investigation of the phospholipase gene family, examining their chromosomal distribution and the characteristics of both their genes and proteins. Using comprehensive and high-quality genome sequences and database resources, they identified 44 genes in the phospholipase gene family from MilletDB. These genes are distributed across the seven chromosomes, with chromosome 1 having the highest count. A detailed expression analysis of these genes was conducted, covering key growth stages and identifying cis-regulatory elements from their putative promoter sequences. The widespread expression of phospholipase genes in all six tissues of two pearl millet genotypes suggests developmental regulation. This aligns with previous findings showing differential transcriptional regulation of PL genes during various growth and developmental stages in other plants.
Genes with overlapping expression patterns across multiple tissues, as well as those specifically expressed in grains, were identified. The expression data were also analyzed between two contrasting genotypes for rancidity, ICMB-863 and ICMB-95222. Five of the 30 PL genes demonstrating expression in grains across both genotypes studied, were grain specific. Within this subset of five genes, comparatively high expression levels were observed for PgPLD-alpha1-1 and PgPLD-alpha1-5 in ICMB-863, a high rancid pearl millet genotype. Targeting grain-specific genes holds promise for enhancing grain quality parameters of pearl millet, including addressing rancidity issues through genetic or genomic engineering approaches.
The study also identified 19 PgPLA members, including 14 PgPLA1 and 5 PgPLA2, with no patatin-like PgPLAs detected. Among these, two PgPLA1-DAD1 isoforms were identified. DAD1 is an sn1-specific acyl-hydrolase involved in key cellular processes such as seed germination, seedling establishment, and tissue growth. PLA1 transcripts are present in almost all plant organs, with different isoforms showing varying degrees of tissue specificity, consistent with the study’s findings. All PgPLA1 members contain multiple stress-responsive cis-elements, indicating their role in spatiotemporal and stress-related processes. Additionally, MeJa-responsive cis-elements were identified in ten of the 14 PgPLA1 putative promoters, aligning with previous studies on PLA1 involvement in jasmonic acid responsiveness. In contrast, PLA2s are secreted phospholipases with a smaller and less complex protein array compared to PLA1s. While only four PLA2 isoforms have been identified in Arabidopsis and rice, five PgPLA2 isoforms were found in pearl millet. Notably, transcripts of PgPLA2-3b were present in all six tissues examined from both genotypes of pearl millet. PLA2 enzymes are also involved in plant growth and defense mechanisms against stresses.
The study highlights the role of phospholipases in grain quality, making it a promising candidate to understand the genetics of this vital trait in a climate resilient crop. Additionally, our recent study using non-targeted metabolite profiling of contrasting pearl millet genotypes showing flour rancidity also suggests these genes as one of the key targets in regulating grain quality ( https://doi.org/10.3390/ijms252111583). – Tyagi
SorghumBase examples:
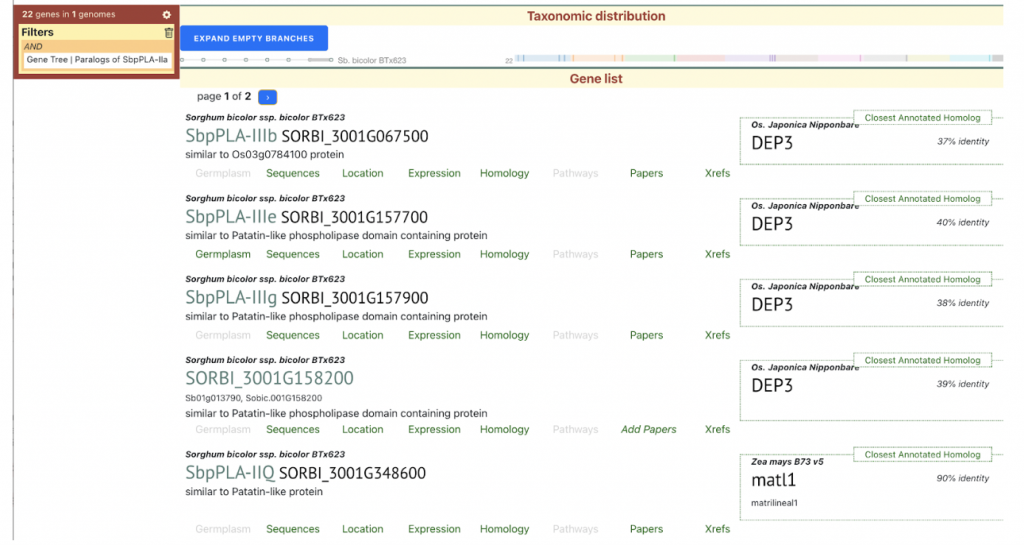
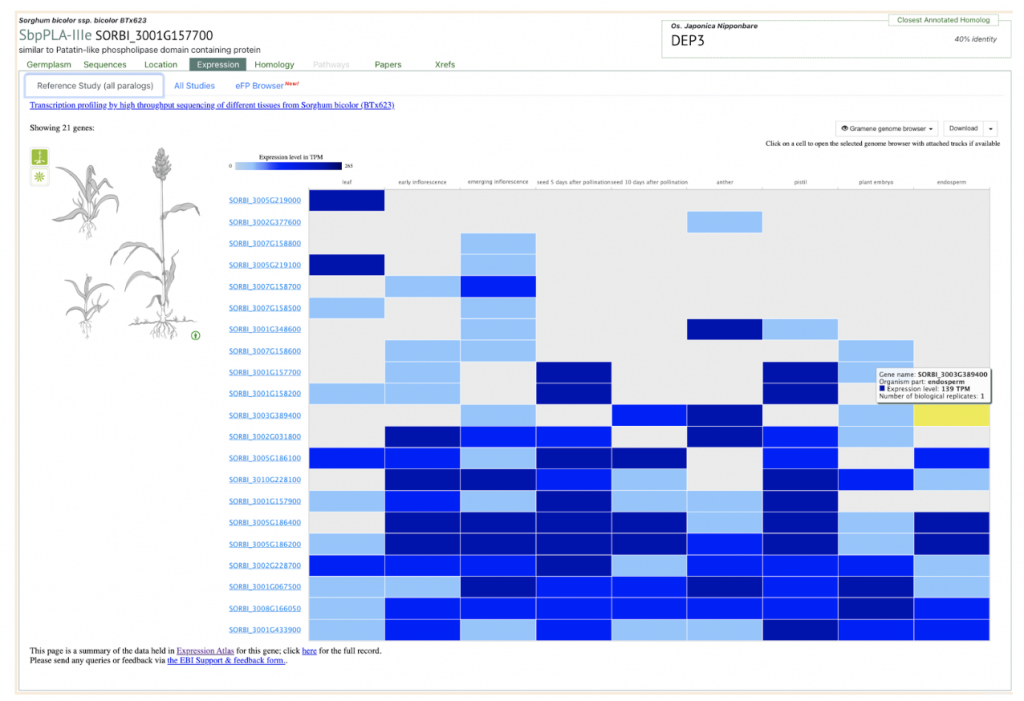
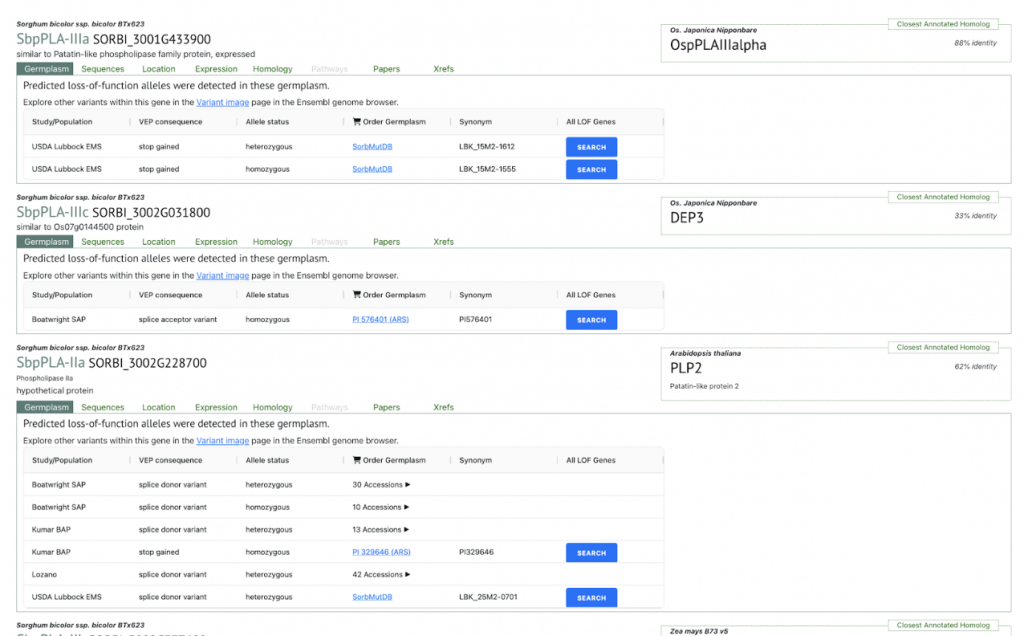
Reference:
Moin M, Bommineni PR, Tyagi W. Exploration of the pearl millet phospholipase gene family to identify potential candidates for grain quality traits. BMC Genomics. 2024 Jun 10;25(1):581. PMID: 38858648. doi: 10.1186/s12864-024-10504-x. Read more
Related Project Websites:
- The Cell, Molecular Biology, and Trait Engineering team page at ICRISAT: https://www.icrisat.org/research/cell-molecular-biology-and-genetic-engineering/about
