Plants are frequently exposed to both biotic and abiotic stressors that threaten their growth, requiring the development of two primary immune strategies: PAMP-triggered immunity (PTI) and effector-triggered immunity (ETI). PTI is initiated when pattern recognition receptors (PRRs) recognize pathogen-associated molecular patterns (PAMPs), while ETI is driven by intracellular nucleotide-binding leucine-rich repeat receptors (NLRs) that detect pathogen effectors. NLRs are divided into coiled-coil (CNL), Toll/interleukin-1 (TNL), and resistance to powdery mildew 8-like (RNL) classes. CNLs are present in both monocots and dicots, while typical TNLs are absent in monocots. NLRs play a crucial role in plant defense by activating ETI and inducing hypersensitive responses (HR). One notable CNL protein, ZAR1, forms a pentameric resistosome that triggers calcium influx and cell death. A mutation in the sorghum NLR protein, SbYR1, demonstrated that it not only confers resistance to sorghum head smut disease but also impacts plant growth by regulating hormone synthesis and flavonoid metabolism.
Researchers from Liaoning Academy of Agricultural Sciences, Shenyang Agricultural University and Liaodong University generated the sorghum sbyr1 mutant via EMS mutagenesis, leading to significant reductions in plant height, leaf length, dry weight, and reproductive traits, such as seed setting rate and grain weight, compared to wild-type BTx623. Physiological changes included reduced chlorophyll content and increased levels of anthocyanins, soluble sugars, and starch, indicating that sbyr1 affects growth through alterations in secondary metabolism. The mutant exhibited enhanced resistance to head smut disease, likely linked to its mutation in the SbYR1 NLR protein, a known pathogenesis-related protein involved in plant immunity. Transcriptomic and metabolomic analyses showed that flavonoid metabolism and genes related to abiotic and biotic stress responses were significantly impacted in sbyr1, suggesting that the mutation may enhance stress tolerance by influencing flavonoid synthesis and hormone signaling pathways such as those involving ABA and JA.
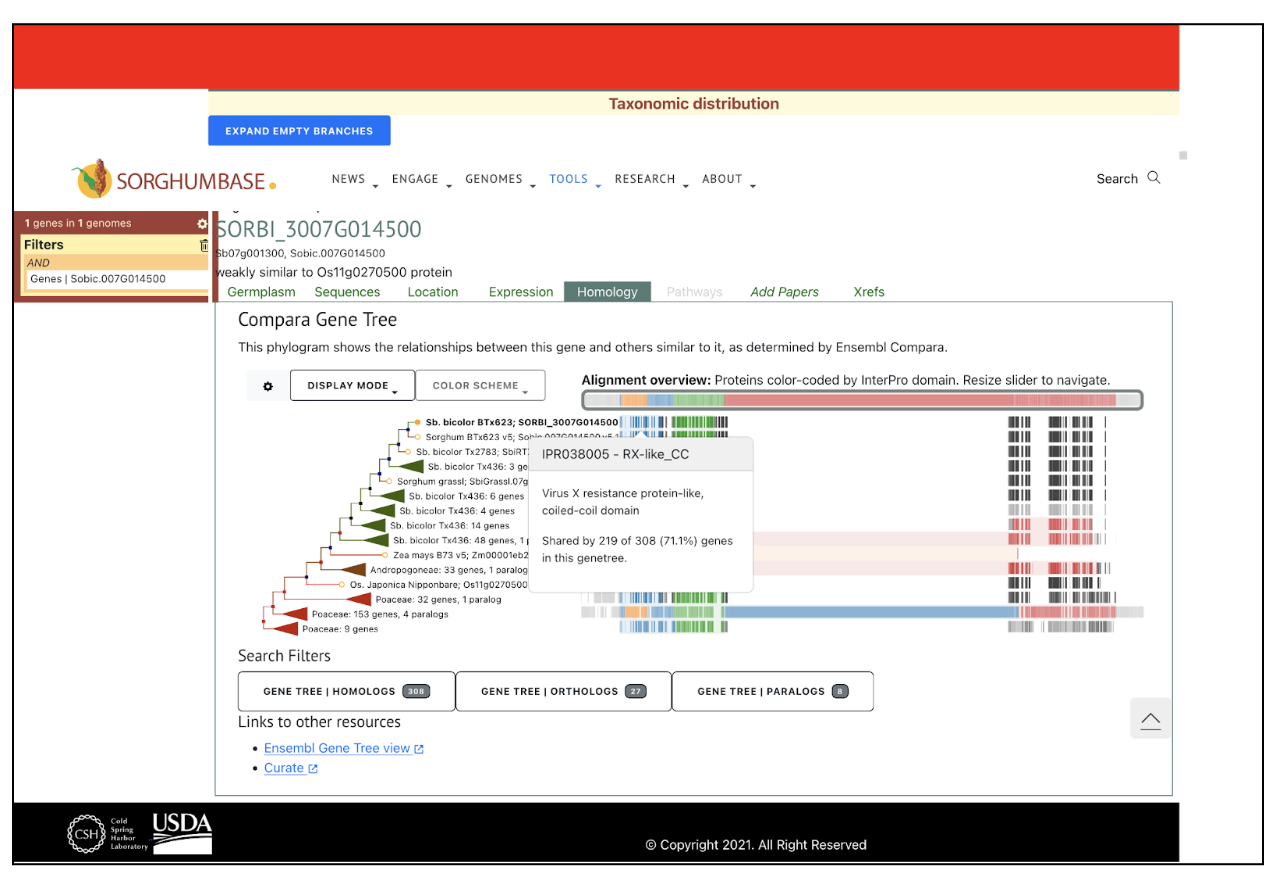
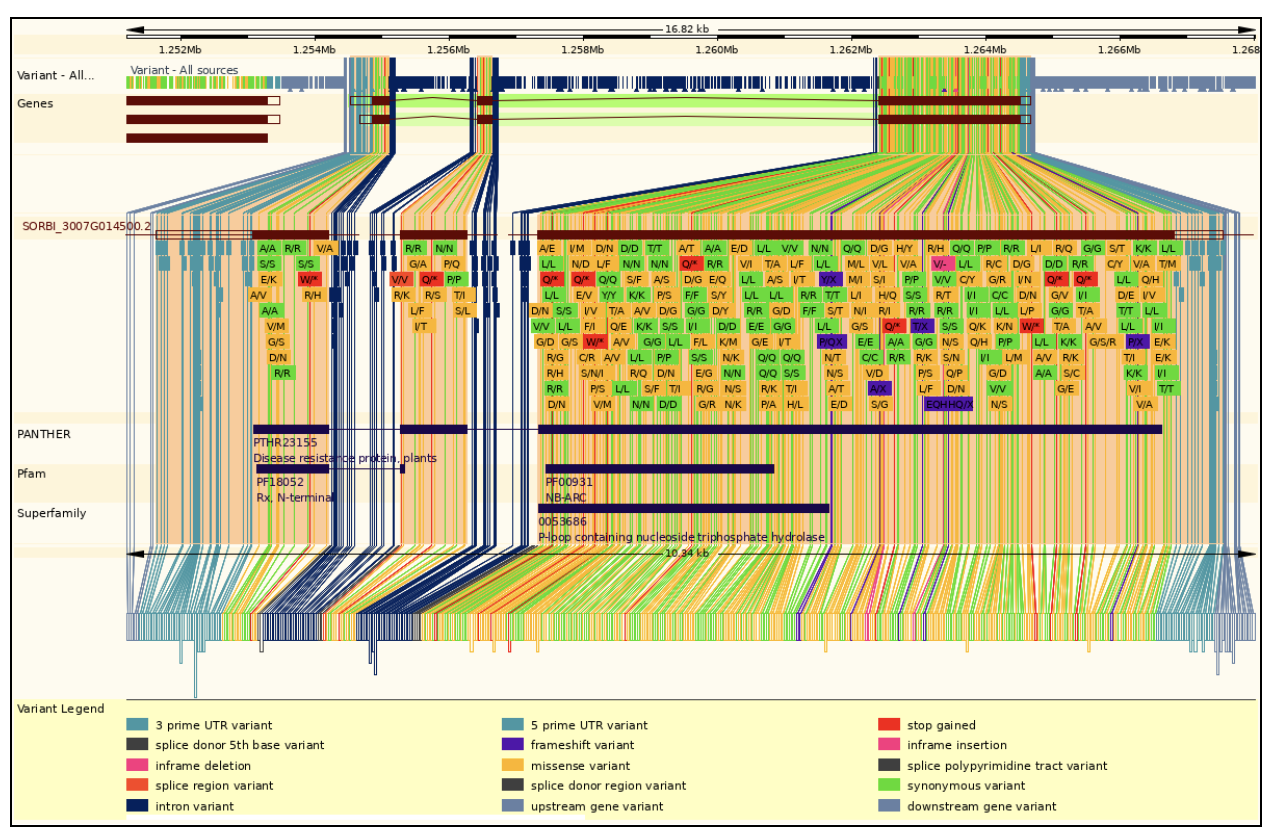
SorghumBase examples:
The SbYR1 (Sobic.007G014500) gene, identified in this study, is a key NLR (nucleotide-binding leucine-rich repeat) receptor gene in Sorghum bicolor with important roles in both growth regulation and stress tolerance. A Thr4Met mutation in its coiled-coil (CC) domain enhances resistance to head smut disease but also causes stunted growth and yellow ripple leaves due to elevated levels of jasmonic acid (JA) and abscisic acid (ABA), which suppress growth. Transcriptome and metabolome analyses show that this mutation influences secondary metabolic pathways, particularly flavonoid biosynthesis, and upregulates pathogenesis-related (PR) proteins, strengthening the plant’s immune response. However, the mutation leads to a trade-off between growth and stress tolerance, offering potential for breeding stress-resistant sorghum varieties with careful management of growth impacts.
Reference:
Li D, Zhu Z, Qu K, Li J, Ma D, Lu X. A coiled-coil domain mutation in the NLR receptor SbYR1 coordinates plant growth and stress tolerance in sorghum. Plant Sci. 2024 Sep 18:112246. PMID: 39304072. doi: 10.1016/j.plantsci.2024.112246. Read more