Research reveals that drought stress disrupts photosynthesis by down-regulating key genes in C3 (wheat) and C4 (sorghum) plants, with wheat exhibiting greater susceptibility, highlighting potential genetic targets for improving drought tolerance in crops.
Keywords: C3, C4, Drought, Meta-analysis, Photosynthesis
Drought stress is one of the most severe environmental factors affecting plant productivity, primarily by inhibiting photosynthesis. This inhibition occurs due to stomatal closure, which limits CO2 intake, reducing the efficiency of the photosynthetic process. Plants have evolved three photosynthetic pathways—C3, C4, and CAM—each with varying degrees of drought tolerance. C4 plants, such as sorghum, demonstrate higher water use efficiency than C3 plants like wheat due to their ability to concentrate CO2 in bundle sheath cells. Scientists from Shahrekord University conducted a transcriptome meta-analysis to investigate the effects of drought stress on photosynthesis-related gene expression in wheat and sorghum. The results revealed a significant down-regulation of genes encoding light-harvesting Chl a/b-binding proteins and ATP synthase subunits, with wheat exhibiting a greater reduction than sorghum. Additionally, genes involved in PSI and PSII function were down-regulated in wheat, leading to disrupted electron transport, increased ROS accumulation and greater photodamage under drought conditions.
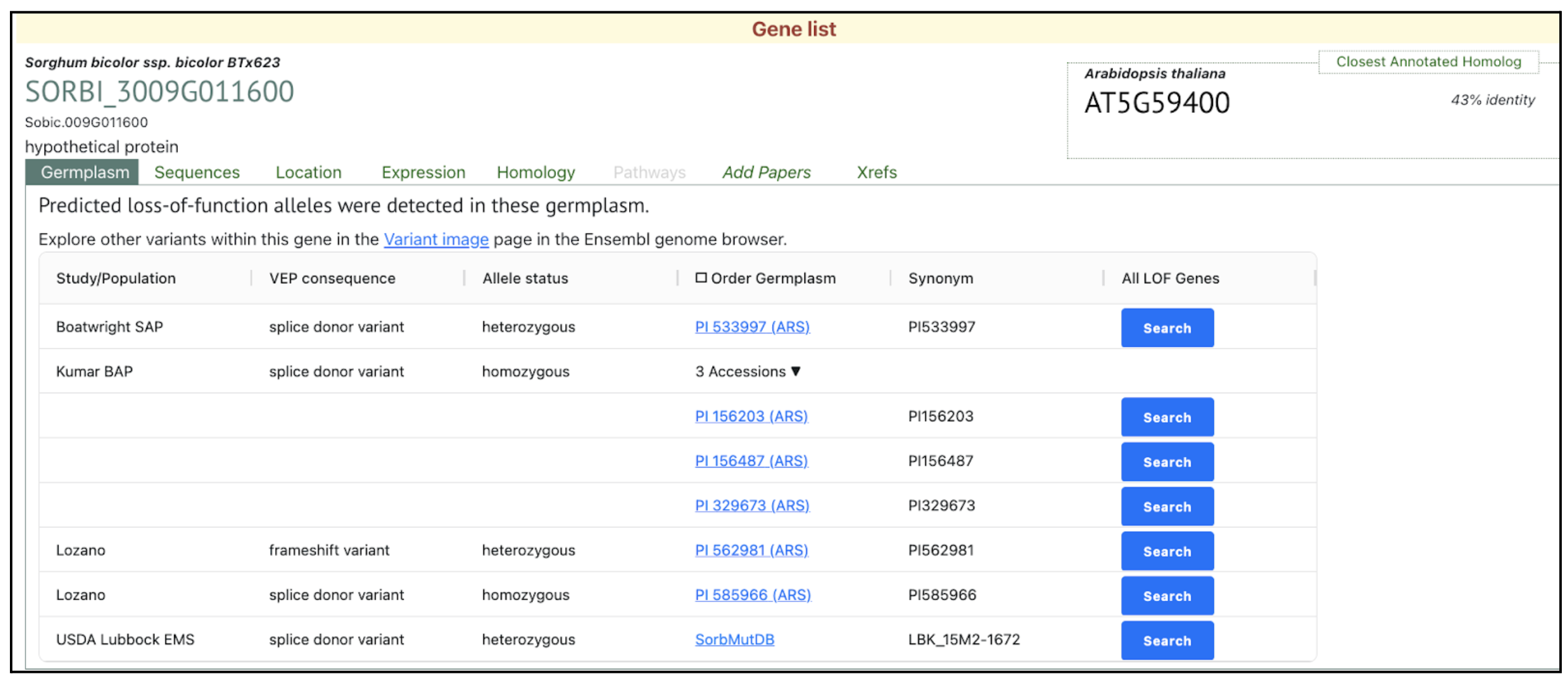
Sorghum exhibited an upregulation of the PGRL1 gene, which is associated with cyclic electron flow (CEF), a protective mechanism against photoinhibition. Conversely, wheat showed a decrease in CEF-related gene expression, indicating a reduced capacity to mitigate oxidative stress. Interestingly, wheat responded to drought by upregulating genes related to C4 photosynthesis, potentially enhancing NADPH production for antioxidant defense and osmotic adjustment. The study also found that drought stress altered chlorophyll metabolism, with wheat exhibiting more extensive changes, possibly due to its greater susceptibility to oxidative damage. These findings highlight key genetic responses to drought stress in C3 and C4 plants and suggest that engineering C4 traits in C3 crops could improve drought tolerance. This research provides valuable insights for developing drought-resistant crop varieties, ultimately enhancing agricultural productivity and food security.
SorghumBase examples:
This study has demonstrated the effects of drought stress in common wheat (Triticum aestivum) and sorghum (Sorghum bicolor), as representatives of C3 and C4 plants. They have investigated the photosynthesis-related genes involved in the response to drought stress using a meta-analysis approach. This paper highlights two important pathways for cyclic electron flow (CEF) that occur in plants. The first pathway involves the proton gradient regulation 5 (PGR5)/PGR5-like 1 (PGRL1) protein and the second pathway involves the chloroplast NADH dehydrogenase-like complex (NDH) components. We have selected PGRL1 gene (SORBI_3009G011600) which was upregulated in sorghum.
Karami S, Shiran B, Ravash R. Molecular investigation of how drought stress affects chlorophyll metabolism and photosynthesis in leaves of C3 and C4 plant species: A transcriptome meta-analysis. Heliyon. 2025 Jan 29;11(3):e42368. PMID: 39981367. doi: 10.1016/j.heliyon.2025.e42368. Read more
Related Project Websites:
- Dr Ravash’s page at Shahrekord University: https://sku.ac.ir/en/DepartmentGroup/ProfessorForm.aspx?ID=2347#