This study identifies and validates new stable dw3 dwarfing alleles using sorghum pangenome data to improve genetic diversity and eliminate height revertants in U.S. grain sorghum breeding.
Keywords: haplotype-based markers, sorghum NAM, sorghum allele mining, stable height, trait improvement
This study exemplifies how pangenome-enabled allele discovery can overcome the genetic bottlenecks imposed by domestication and selective breeding, offering stable alternatives to legacy alleles like dw3-ref and opening new avenues for enhancing sorghum’s agronomic performance and resilience in the face of climate change. – Felderhoff
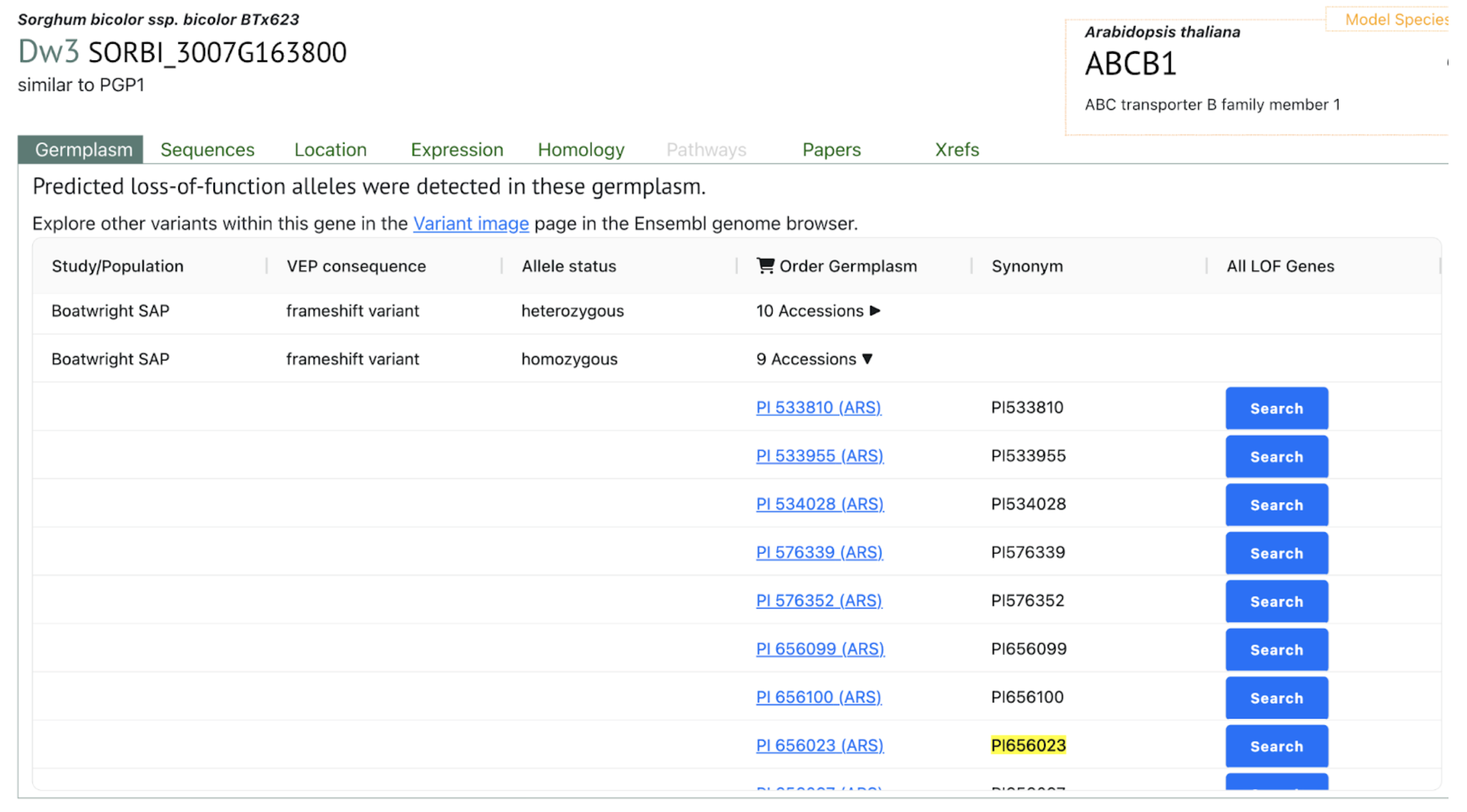
Domestication and selective breeding for agronomic traits like reduced height and photoperiod insensitivity in sorghum have led to genetic sweeps that reduced diversity around key loci, especially those controlling plant height and flowering time. To address this, the U.S. sorghum conversion (SC) programs crossed exotic sorghum lines with a temperate elite donor, BTx406, to develop semi-dwarf, photoperiod-insensitive varieties suitable for mechanized harvesting. However, despite broadening genetic backgrounds, selection for specific dwarfing (Dw1–Dw4) and maturity (Ma1–Ma6) alleles fixed large chromosomal regions, notably around the Dw3 locus. The widely used dw3-ref allele, although effective in reducing plant height, is unstable and produces revertants due to an 882-bp tandem duplication, increasing seed production costs. In this study, researchers from Kansas State University and Colorado State University leveraged pangenome and resequencing data from over 2,000 sorghum lines to identify novel dw3 loss-of-function alleles that avoid the instability of dw3-ref and can be introgressed into elite U.S. lines using marker-assisted selection.
The team identified and validated seven complete loss-of-function dw3 alleles that suppressed revertant production in field trials, including dw3-sd6, which lacks the tandem duplication. Using high-resolution genomic resources like the sorghum pangenome, they developed haplotype-based KASP markers to guide precise introgression of these stable alleles into breeding programs. Unlike dw1 and dw2, where only one dwarfing allele exists, the dw3 locus shows greater natural variation, providing opportunities to expand genetic diversity while maintaining agronomic performance. Furthermore, the study underscores the potential of combining pangenome-enabled allele discovery with CRISPR and mutagenesis approaches to generate novel alleles at fixed loci, ultimately enhancing the genetic resilience and productivity of sorghum in changing climates.
SorghumBase examples:

Reference:
Marla SR, Olatoye M, Davis M, Otchere V, Sexton-Bowser S, Morris GP, Feldherhoff T. Mining sorghum pangenome enabled identification of new dw3 alleles for breeding stable-dwarfing hybrids. G3 (Bethesda). 2025 Mar 12:jkaf054. PMID: 40071298. doi: 10.1093/g3journal/jkaf054. Read more
Related Project Websites:
- Terry Felderhoff’s page at Kansa State University: https://www.agronomy.k-state.edu/about/people/faculty/felderhoff-terry/
- Morris Lab at Colorado State University: https://www.morrislab.org/home
- Sarah Sexton-Bowser’s page at Kansa State University: https://www.agronomy.k-state.edu/about/people/faculty/sexton-bowser-sarah/
- Center for Sorghum Improvement (CSI): https://www.csisorghum.org/