Mishra et al. used multi-omics analysis in sorghum to reveal conserved gene networks and regulatory mechanisms underlying iron and zinc homeostasis, linking root uptake and leaf chloroplast function under micronutrient stress.
Keywords: Differential expression, gene regulatory network, iron, sorghum, zinc
Our multi-omics approach revealed how sorghum dynamically regulates iron and zinc homeostasis through interconnected gene networks that span root and shoot tissues. These insights highlight the crop’s remarkable capacity to adapt to fluctuating nutrient and water availability—knowledge that is critical for guiding future efforts to breed sorghum varieties better suited to challenging environments. – Paape
Sorghum (Sorghum bicolor) is a drought-tolerant C₄ crop widely cultivated for grain and bioenergy purposes across diverse and often challenging environments. Essential micronutrients such as iron (Fe) and zinc (Zn) are critical for sorghum growth and development, but their deficiency or excess can lead to physiological stress. Using a multi-omics approach, including ionomics and transcriptomics, scientists from Texas A&M University, Brookhaven National Laboratory, Cold Spring Harbor Laboratory, Argonne National Laboratory, Lawrence Berkeley National Laboratory and the USDA-ARS conducted a time-series experiment on the reference genotype Btx623 under conditions of both Fe- and Zn-deficient and excess. The study aimed to profile ion accumulation patterns and tissue-specific transcriptomic responses over a 3-week period. Transcriptome analyses revealed upregulation of key transport and chelation genes (e.g., SbSAMS, SbNAS4, SbIDS3, SbOPT3, SbNRAMP2, SbZIP10) in both root and leaf tissues, indicating systemic regulation of micronutrient transport. Transcriptomic cross-talk between Fe and Zn responses suggests tightly coordinated mechanisms underlying micronutrient homeostasis, potentially shaped by seasonal variability in water availability, such as early-season flooding and late-season drought. This supports the hypothesis that sorghum may switch between Strategy I and Strategy II iron acquisition pathways depending on environmental conditions.
Gene regulatory network (GRN) analysis further identified conserved hub genes (SbPYE, SbBTS, SbFIT) associated with both Fe limitation and Zn excess responses in roots and leaves. These hub genes were also implicated in chloroplast function, including Fe–S cluster assembly, photosynthesis, and Reactive oxygen species (ROS) detoxification, highlighting their central roles in coordinating below ground nutrient uptake and above ground metabolic processes. This dataset provides a valuable foundation for functional genomics and crop improvement efforts aimed at enhancing nutrient use efficiency, particularly in environments with variable soil and climatic conditions.
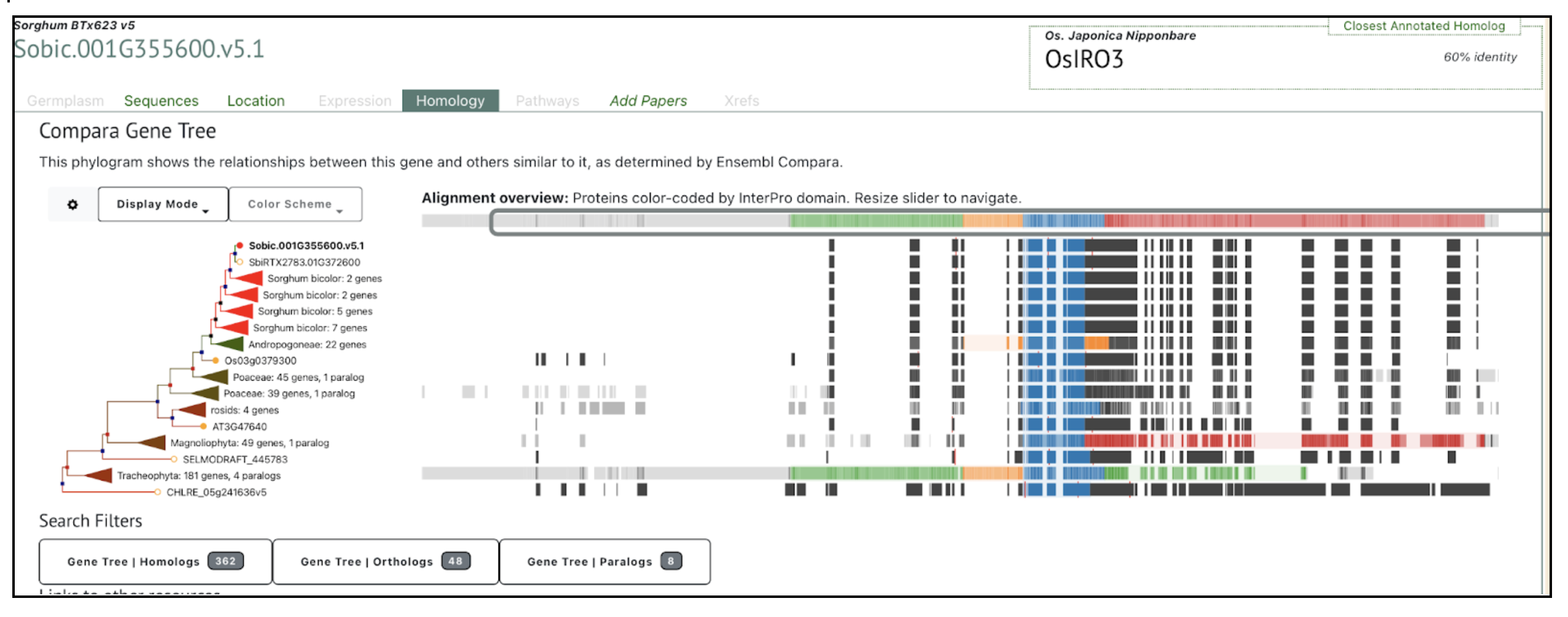
SorghumBase example:
Mishra A, Bhat A, Kumari S, Sharma R, Braynen J, Tadesse D, El Alaoui S, Seaver S, Grosjean N, Ware D, Xie M, Paape T. Time-series multi-omics analysis of micronutrient stress in Sorghum bicolor reveals iron and zinc crosstalk and regulatory network conservation. Plant Biol (Stuttg). 2025 May 22. PMID: 40402192. doi: 10.1111/plb.70038. Read more
Related Project Websites:
- Tim Paape’s page at USDA-ARS and Texas A&M University: https://iha.tamu.edu/person/tim-paape/
- Responsive Agricultural Food Systems Research Unit at the USDA: https://www.ars.usda.gov/plains-area/college-station-texas-rafsru/responsive-agricultural-food-systems-research-unit/
- Meng Xie’s page at Brookhaven National Lab: https://www.bnl.gov/staff/mxie
- Doreen Ware’s page at Cold Spring Harbor Lab: https://www.cshl.edu/research/faculty-staff/doreen-ware/