Sorghum (Sorghum bicolor L. Moench) is a crucial cereal crop, particularly in sub-Saharan Africa, serving as a staple food and providing energy, protein, and other essential nutrients. Despite its importance, sorghum productivity faces challenges from various biotic and abiotic stresses, with drought being a significant limiting factor. Root system architecture (RSA) plays a critical role in drought adaptation, making it essential to understand the genetic basis of RSA traits for sorghum improvement.
Scientists from Haramaya University, Kansas State University, the Ethiopia Agricultural Transformation Institute, the USDA-ARS, the Ethiopian Institute of Agricultural Research (EIAR) and Jimma University investigated the genetic variability and genomic regions associated with RSA traits in a panel of 274 Ethiopian sorghum accessions to enhance sorghum’s tolerance to environmental stress. Utilizing multiple methodologies, including multi-locus genome-wide association study (ML-GWAS) with 265,944 high-quality single nucleotide polymorphism markers, the study identifies 17 reliable quantitative trait nucleotides (QTNs) significantly linked to root angle, number, length, and dry weight. These QTNs, located on chromosomes SBI-01, SBI-02, and SBI-05, include both previously identified and novel genomic regions, with 118 genes colocated with the QTNs. Additionally, the study highlights five intragenic QTNs, three of which are associated with genes responsible for plant growth hormone-induced RSA, offering potential targets for further genetic exploration to enhance sorghum’s root development in drought-prone regions. The findings provide valuable insights into enhancing sorghum’s drought tolerance through genetic improvement of its root system architecture.
A complex dance of survival: Seedling root angle and length are key elements in sorghum’s drought adaptation strategy, working in tandem with other factors like leaf morphology and physiological responses to optimize water uptake and minimize stress. – Menamo
SorghumBase examples:
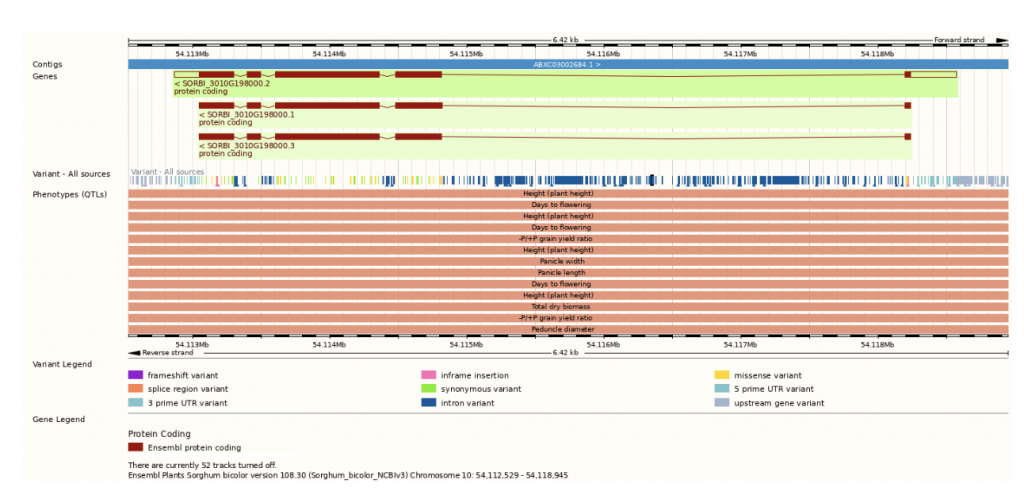
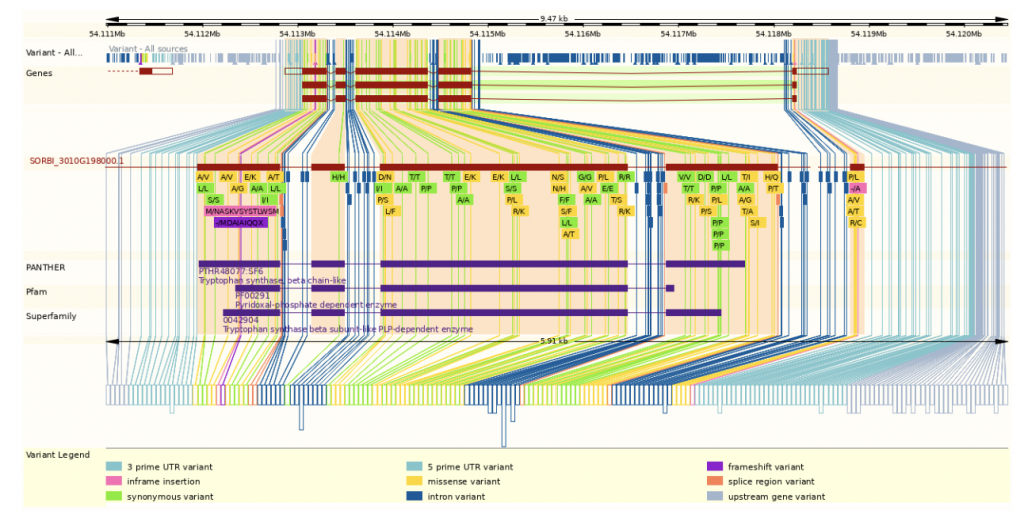
Reference:
Elias M, Chere D, Lule D, Serba D, Tirfessa A, Gelmesa D, Tesso T, Bantte K, Menamo TM. Multi-locus genome-wide association study reveal genomic regions underlying root system architecture traits in Ethiopian sorghum germplasm. Plant Genome. 2024 Feb 15:e20436. PMID: 38361379. doi: 10.1002/tpg2.20436. Read more
Related Project Website:
The Plant and Animal Biotechnology Research Center at Jimma University: https://ju.edu.et/plant-and-animal-biotechnology-research-center/
A

B

C

D
