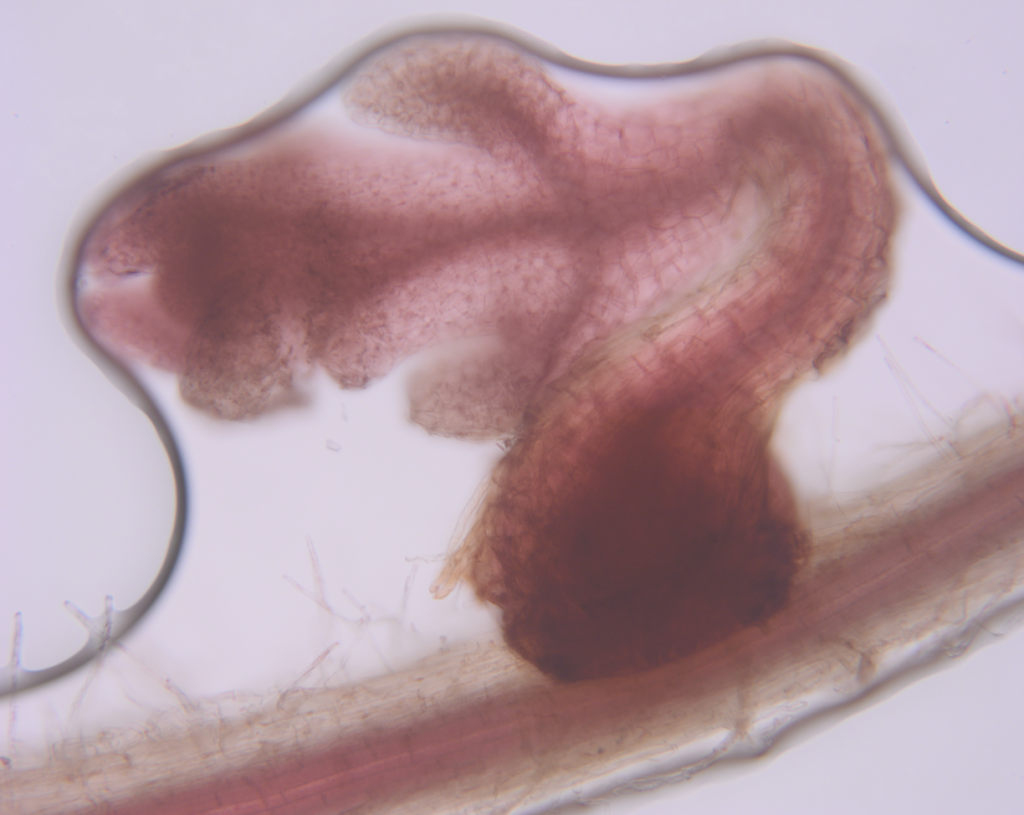
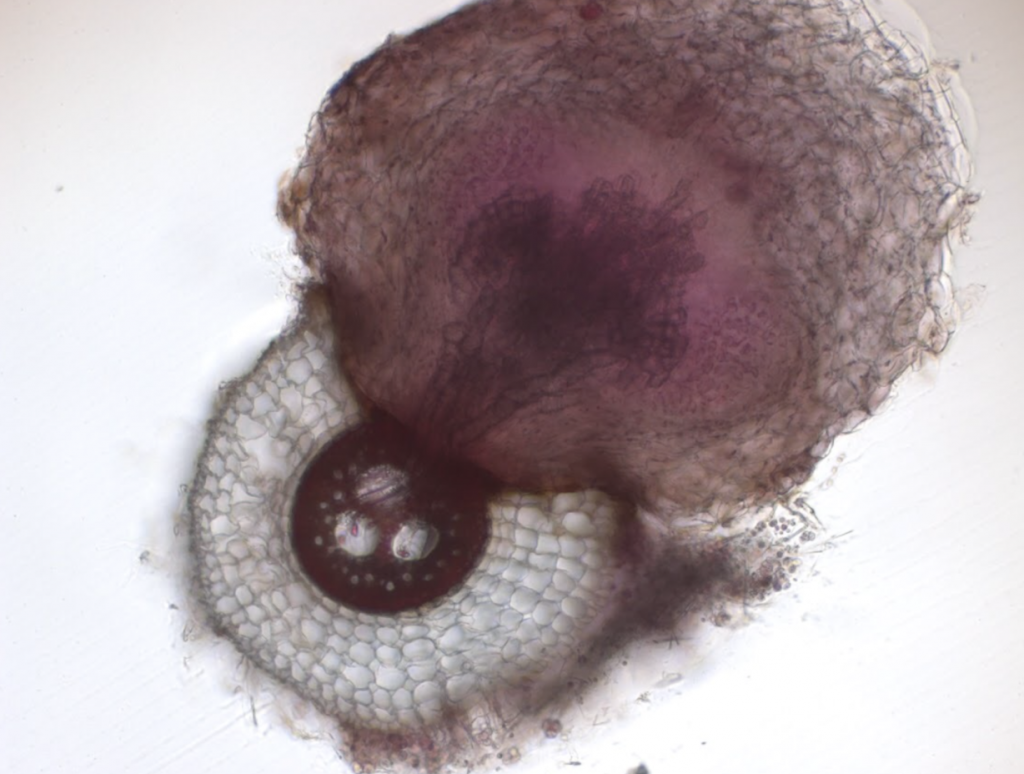
Sorghum bicolor, a vital cereal crop globally, faces significant yield losses due to infestation with the root parasitic weed Striga hermonthica, particularly affecting sub-Saharan Africa’s smallholder farmers. Despite efforts to manage Striga, including chemical control and breeding for resistance, current strategies offer only partial mitigation and are often inaccessible to smallholder farmers. Microbial-based solutions have emerged as promising alternatives due to their potential integration into existing agricultural practices and effectiveness in suppressing various plant diseases. Understanding the soil microbiome’s role in Striga suppression could provide valuable insights into developing sustainable management strategies.
Recent research led by scientists from the University of California Davis along partners from international consortium, indicates that the soil microbiome can suppress Striga infection in sorghum roots by disrupting host-parasite signaling and modulating root cellullar anatomy. The latter included formation of aerenchyma and increased suberin deposition in endodermis. Soil microbiome from Striga-suppressive soil was studied and Pseudomonas strain VK46 was found to reduce haustorium formation in sorghum roots by degrading key haustorium-inducing factors (HIFs) and Arthrobacter strain VK49 was found to induce endodermal suberin deposition. To determine the potential molecular mechanisms that underlie these changes, this group conducted transcriptome profiling of the sorghum root systems at both 2 and 3 weeks post-infection, grown in natural soil (with microbes) and steralized soil (without microbes), and in the presence and absence of Striga. The transcripts of several sorghum orthologs of suberin biosynthetic genes or putative transporters—Sobic.003G368100 (SbASFTa) and Sobic.005G122800 (SbASFTb), Sobic.004G010300 (SbGPAT4/8), Sobic.009G16200 (SbGPAT5/7), and Sobic.001G413700 (SbABCG1/SbABCG2/ABCG6/ABCG20a) were upregulated in natural soil compared to sterilized soil in the presence and absence of Striga. These transcriptome data align with the observed microbiome-mediated changes of host root cellular anatomy, which in turn is correlated with reduced Striga infection.
While these findings provide a promising foundation for developing microbial-based Striga management strategies, further research is needed to validate the efficacy of microbial consortia in field conditions and understand their interaction with host genotypes and environmental factors. Additionally, the study highlights the importance of incorporating comprehensive taxonomic and functional markers in the selection of candidate bacterial isolates for future investigations.
Identified microbes can be used as seed inoculants or soil ammendments. This offers a promise of accessible and affordable Striga managment tool for small-holder sorghum famers in Sub-Saharan Africa. – Kawa
Root suberin and aerenchyma are also known to benefit a plant under drought, thus their application may be extended beyond protections from Striga. – Kawa
SorghumBase examples:
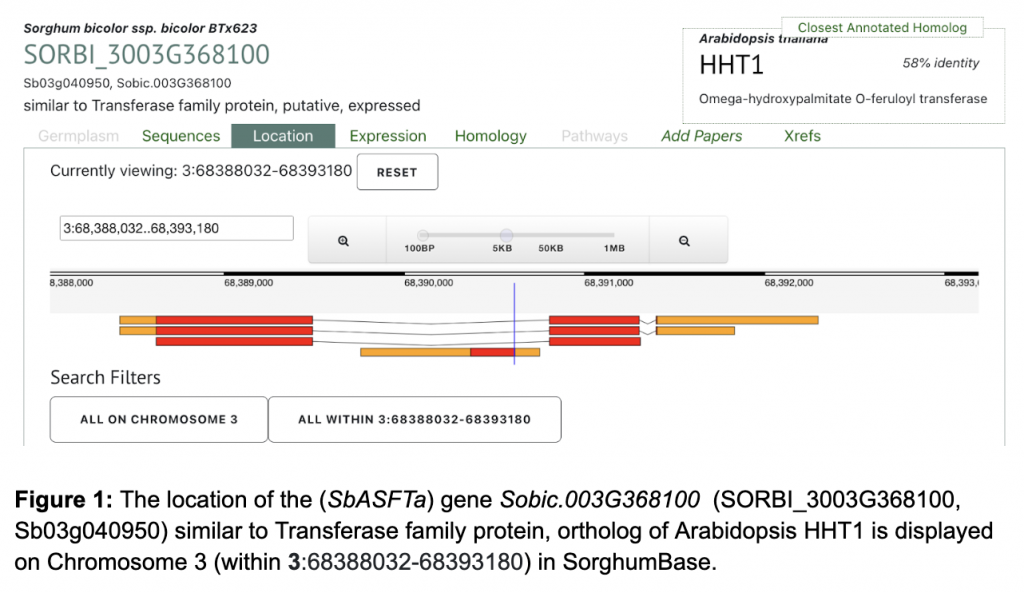
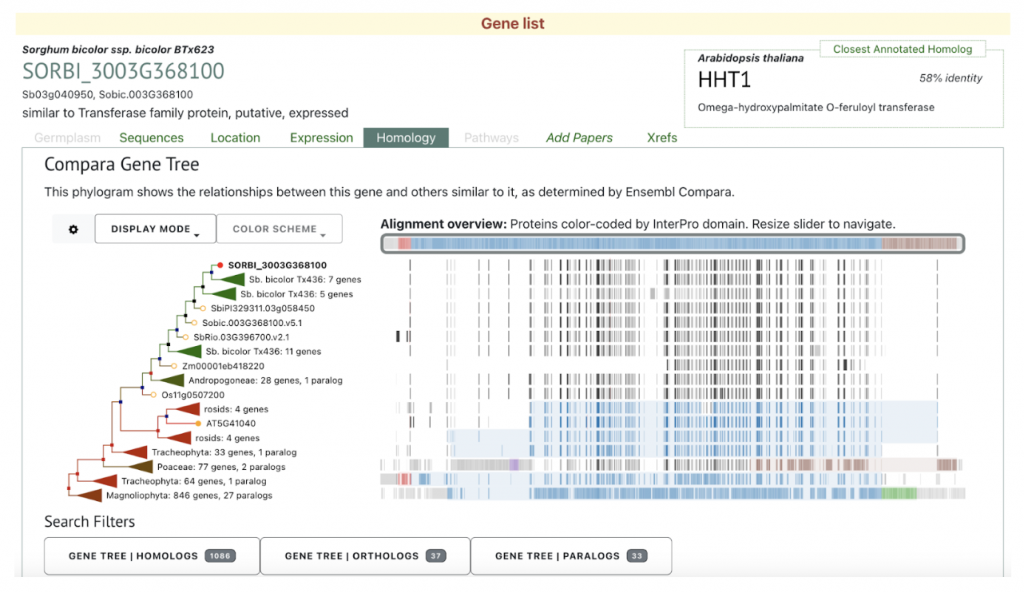
Sobic.003G368100 (SbASFTa) (SORBI_3003G368100) was picked up from the manuscript as an example to explore SorghumBase below:
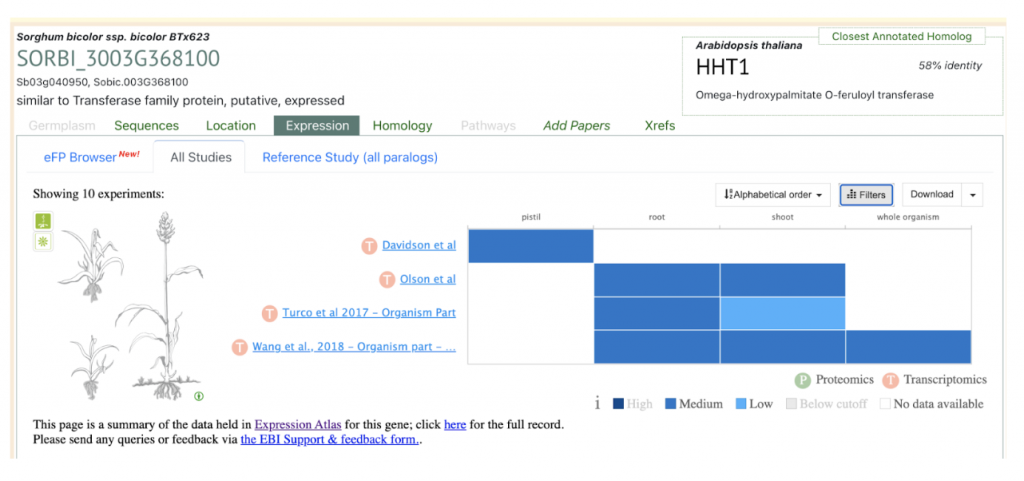
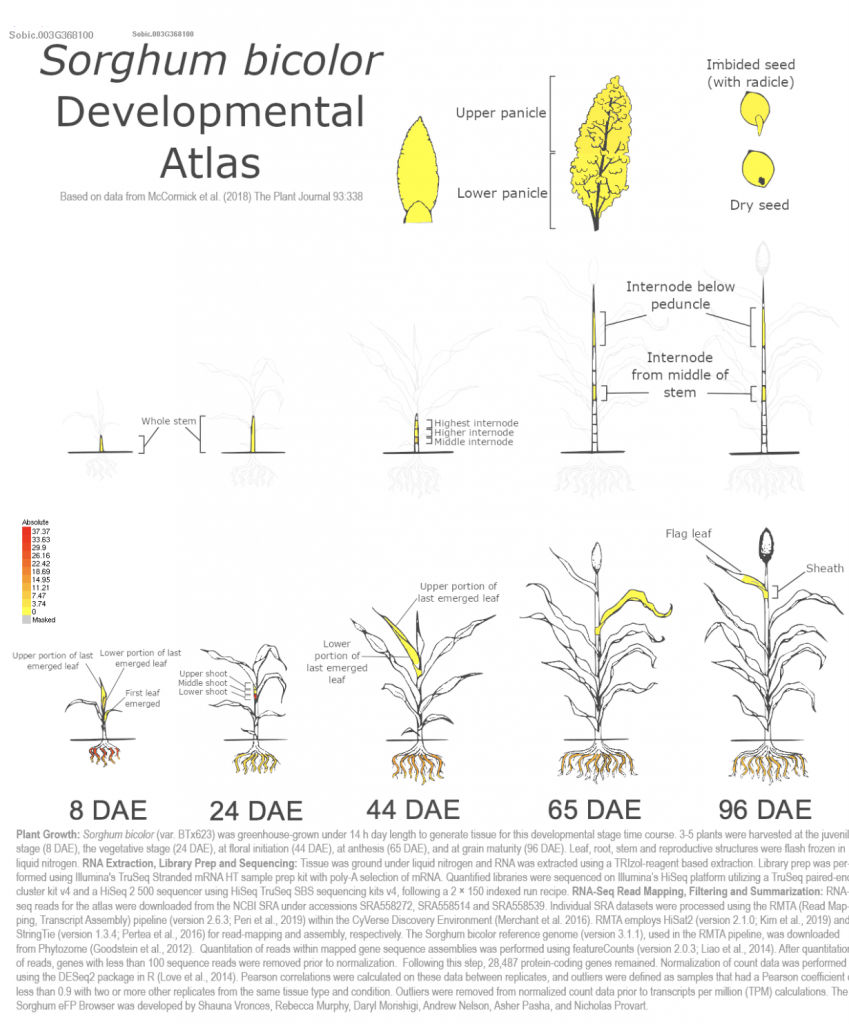
Reference:
Kawa D, Thiombiano B, Shimels MZ, Taylor T, Walmsley A, Vahldick HE, Rybka D, Leite MFA, Musa Z, Bucksch A, Dini-Andreote F, Schilder M, Chen AJ, Daksa J, Etalo DW, Tessema T, Kuramae EE, Raaijmakers JM, Bouwmeester H, Brady SM. The soil microbiome modulates the sorghum root metabolome and cellular traits with a concomitant reduction of Striga infection. Cell Rep. 2024 Mar 26:113971. PMID: 38537644. doi: 10.1016/j.celrep.2024.113971. Read more
Related Project Websites:
- Brady Lab at University of California, Davis: https://www.bradylab.org/
- PROMISE consortium: https://promise.nioo.knaw.nl/en
- Kawa lab at Utrecht University: https://www.uu.nl/staff/DKawa/