Sorghum, a critical crop within the Poaceae family, plays an essential role in agriculture, especially in semi-arid regions where it serves as a staple for over 750 million people. This versatile cereal crop supports human and animal nutrition, ranking as India’s third most significant crop overall. Integral to various cellular functions such as carbon partitioning, cell elongation, defense responses, and plant growth are the enzymatic activities of phospholipases, which hydrolyze phospholipids to produce free fatty acids (FFAs) and other essential compounds. These enzymes, divided into three main groups—PLA, PLC, and PLD—play crucial roles in processes like intracellular signaling, membrane maintenance, and lipid metabolism, with PLA subtypes (pPLA, PLA1, and PLA2) showing specific activities depending on their cleavage positions on phospholipids.
Through extensive analysis of the sorghum genome, researchers from ICRISAT, Osmania University, University of Nottingham and Rothamsted Research identified 32 SbPLA genes, grouped into pPLA, PLA1, and sPLA2 categories, akin to the classifications in Arabidopsis and rice. Subcellular localization studies revealed that these genes are predominantly found in chloroplasts, reflecting similar distribution patterns observed in other plant species. Domain analysis highlighted distinct features such as the ‘patatin’ domain in pPLA, ‘lipase3’ in PLA1, and ‘PA2c’ in sPLA2, marking their functional diversity. Chromosomal mapping showed the genes’ variable distribution across ten chromosomes, excluding chromosomes 6 and 9. Previous research has highlighted the crucial role of the phospholipase gene family in early developmental stages, with many genes showing developmental-specific elements in their promoter regions. Phospholipases are essential for developmental and hormone-specific gene functions and play a significant role in signaling pathways triggered by abiotic stress. The STRING analysis conducted in this study revealed that the SbPLA genes are predominantly involved in lipid degradation, lipid metabolism, and hydrolase activities. SbSDP1-L and SbPLA2a emerged as key regulators, playing significant roles in various processes and overseeing the PLA gene family network. As members of the lipolytic acyl hydrolase family, these proteins are crucial for regulating membrane functions.
Expression profiling suggested that certain SbPLA genes, particularly within the pPLA class, are upregulated during specific developmental stages like root and seedling growth, indicating their potential involvement in seed development and root elongation. Additionally, pollen-specific PLA genes point towards their role in haploid induction, crucial for breeding processes. These findings provide a foundational understanding for further functional and molecular characterization of PLA genes in sorghum, paving the way for advanced studies on their roles in stress responses, growth regulation, and lipid metabolism.
SorghumBase examples:
This study identifies and characterizes 32 phospholipase A (PLA) genes in Sorghum bicolor. We selected the gene SbpPLA-1(Sobic.002G320900) from this published study as the candidate gene to explore SorghumBase (https://sorghumbase.org/).
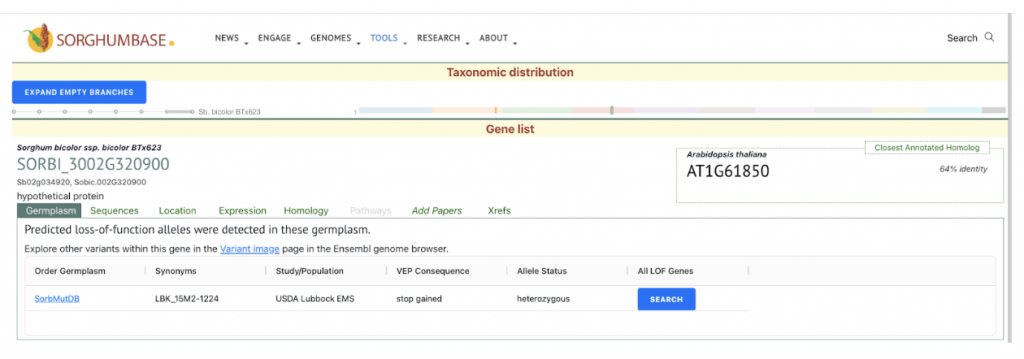
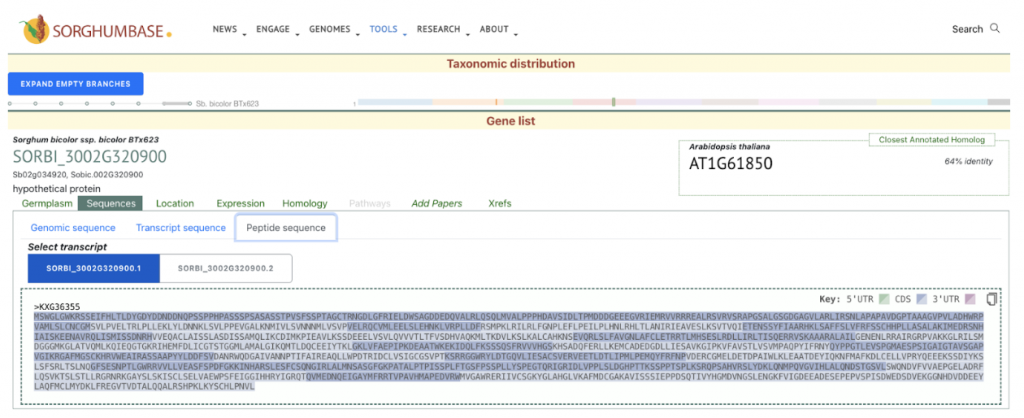
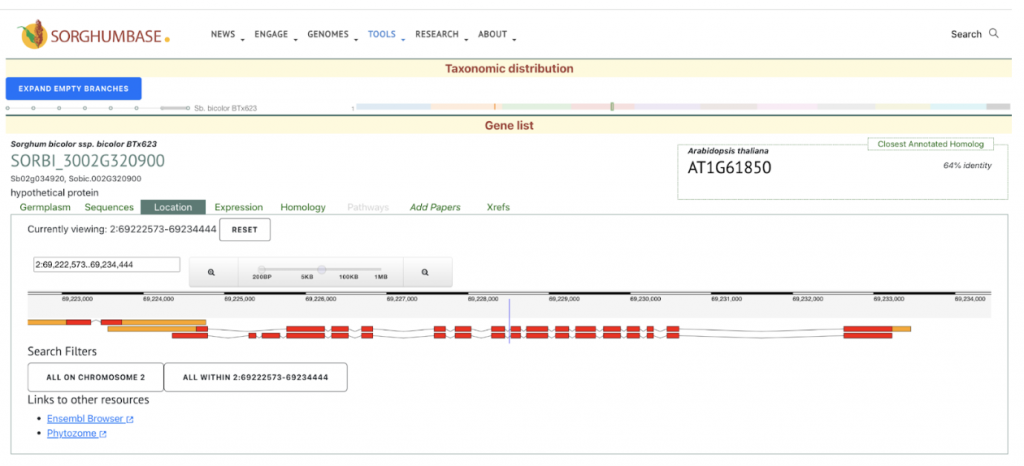
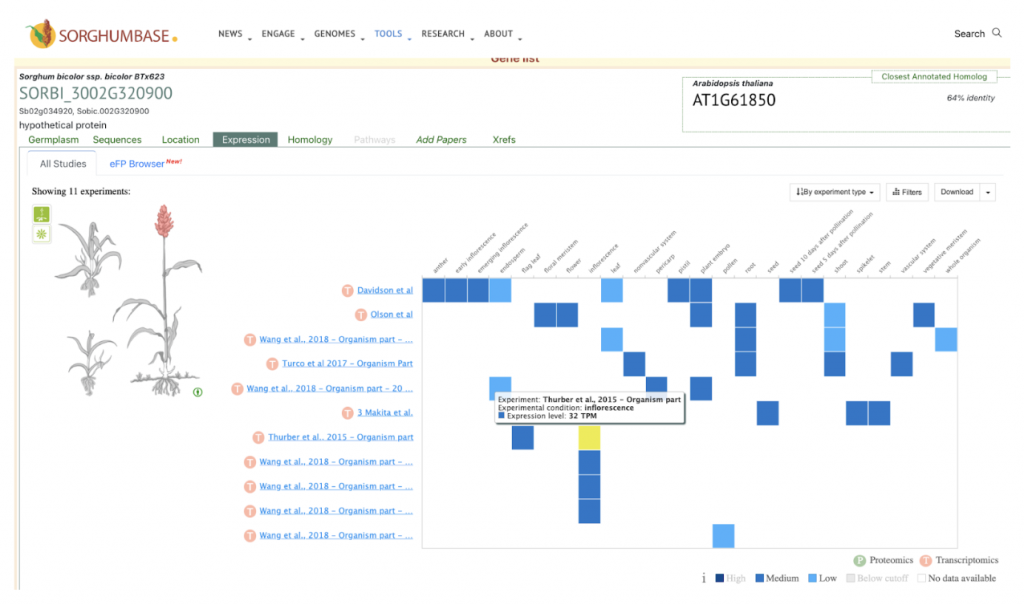
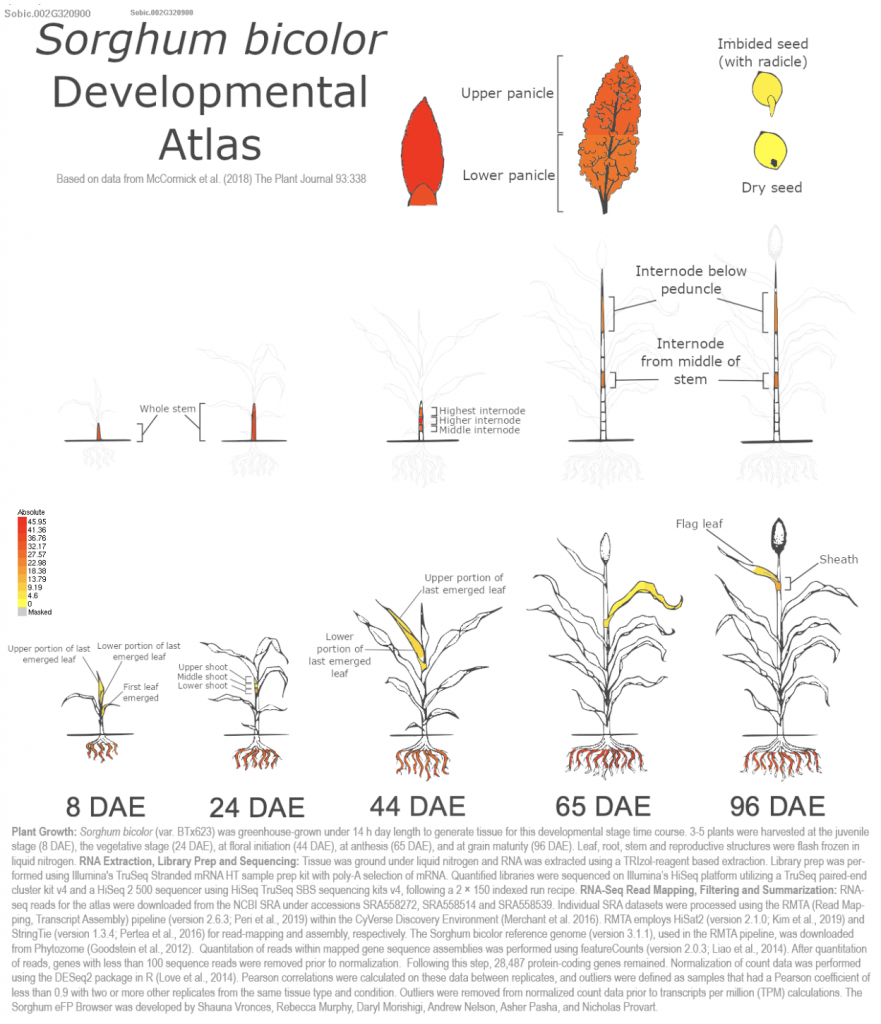
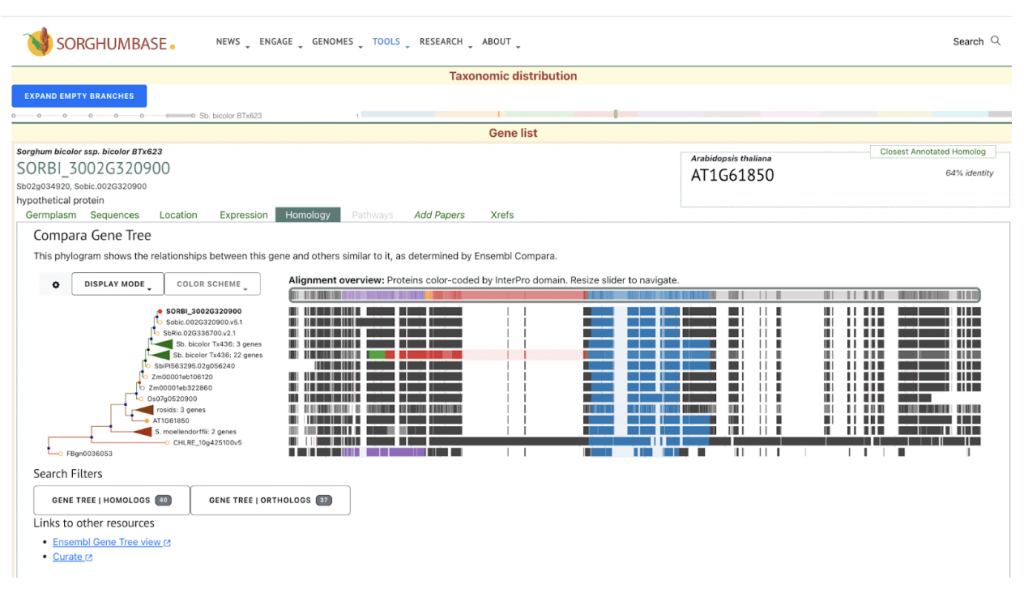
Sapara VJ, Shankhapal AR, Reddy PS. Genome-wide screening and characterization of phospholipase A (PLA)-like genes in sorghum (Sorghum bicolor L.). Planta. 2024 Jun 26;260(2):35. PMID: 38922509. doi: 10.1007/s00425-024-04467-2. Read more
Related Project Websites:
- Dr. Reddy’s page at ICRISAT: https://www.icrisat.org/about/team/dr-sudhakar-reddy-palakolanu