Sorghum (Sorghum bicolor), a major cereal crop, is vital for global food security and is well-suited for climate change resilience. Its seeds consist of genetically distinct tissues: a diploid embryo, a triploid endosperm, and diploid maternal tissues, developing through double fertilization. Sorghum seed development spans around 40-45 days, involving three primary stages: early (pre-6 days post-anthesis [dpa]), middle (6–24 dpa), and late (25–35 dpa), with critical starch accumulation starting at 5 dpa. Key enzymes, such as ADP-glucose pyrophosphorylase and starch synthases, play central roles in starch metabolism, while kafirins, the primary storage proteins, comprise most of the endosperm’s protein content. Despite the nutritional significance of sorghum, there has been limited research into the molecular processes governing its seed development, especially in comparison to other cereal crops like maize and rice.
Recent advancements in transcriptomic and metabolomic studies provide valuable insights into sorghum seed development. Researchers from Texas Tech University, the USDA-ARS and University of Nevada-Reno explored the transcriptomic and metabolomic dynamics of sorghum seed development, offering insights into the molecular mechanisms that govern grain quality and yield. Using RNA-Seq and untargeted metabolome profiling, the research reveals how gene expression and metabolite accumulation are coordinated across five developmental stages in the whole seed, embryo, and endosperm. The findings emphasize the role of tissue-specific genes (TS genes) in shaping seed traits, highlighting critical processes like cell wall biosynthesis in the early seed and starch and protein storage in the endosperm. the potential for improving sorghum through genetic and breeding strategies. Integrating these findings with metabolomic data offers a deeper understanding of gene regulation during seed maturation. This knowledge is crucial for enhancing sorghum’s grain quality, yield, and adaptability to environmental stresses.
This project represents an exciting collaboration among sorghum geneticists, breeders, and grain chemists. The data establishes a foundational understanding of sorghum seed development in the reference genome line, providing a baseline for investigating the diversity of sorghum seed development and grain quality. – Jiao
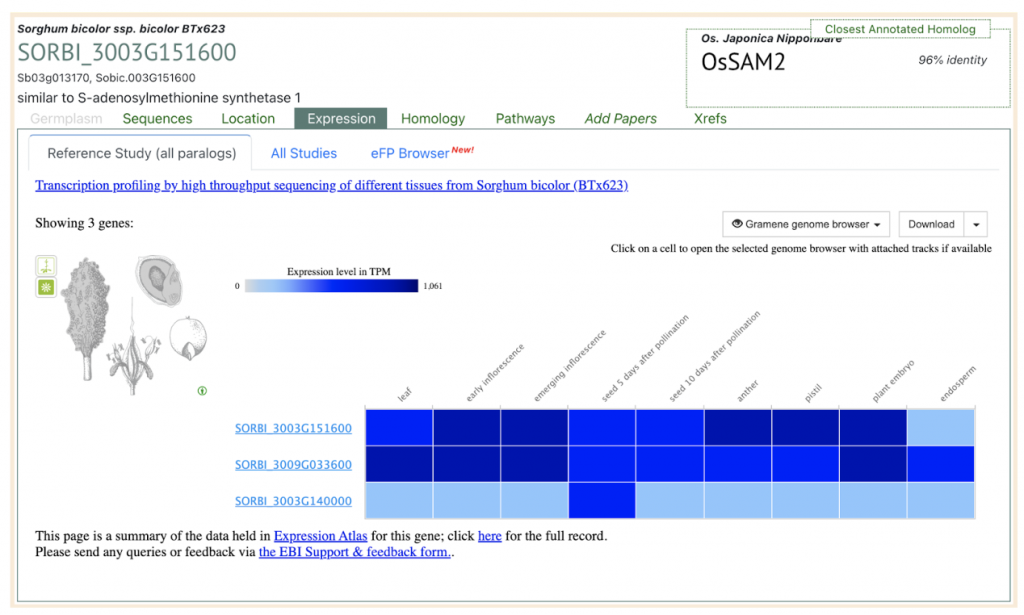
SorghumBase example:
Reference:
Khan A, Tian R, Bean SR, Yerka M, Jiao Y. Transcriptome and metabolome analyses reveal regulatory networks associated with nutrition synthesis in sorghum seeds. Commun Biol. 2024 Jul 10;7(1):841. PMID: 38987396. doi: 10.1038/s42003-024-06525-7. Read more
Related Project Websites:
- Jiao Lab at Texas Tech University: https://www.depts.ttu.edu/igcast/Staff/jiao_lab.php
- Scott Bean’s page at the USDA-ARS: https://www.ars.usda.gov/plains-area/mhk/cgahr/gqsru/people/scott-bean/
- Melinda Yerka’s page at the University of Nevada, Reno: https://www.unr.edu/environmental-sciences-and-health/people/melinda-yerka
