Arid regions face significant challenges in agricultural productivity due to limited soil water availability and high evaporation rates, which create osmotic stress on plants. Traditional crops in these regions require substantial irrigation, but have insufficient yield. Sweet sorghum (Sorghum bicolor), a C4 plant, stands out as a promising alternative. It thrives in harsh conditions, including drought and salinity, while offering high fermentable sugar content, making it a valuable bioenergy and forage crop. Understanding the mechanisms of sweet sorghum’s stress tolerance is crucial for improving agricultural productivity in such environments.
To cope with osmotic stress, plants deploy various adaptive strategies, such as enhancing root systems, minimizing water loss through stomatal closure, and accumulating osmolytes, like soluble sugars and amino acids, for osmotic adjustment. Soluble sugars, such as sucrose, raffinose, and trehalose, are key organic osmolytes that help maintain cell hydration under drought conditions, while also acting as signal molecules and antioxidants. In sweet sorghum, the accumulation of proline, a known osmolyte and antioxidant, plays a crucial role in reactive oxygen species degradation and stress adaptation. Scientists from Northwest A&F University in China investigated the impacts of different concentrations of polyethylene glycol (PEG) on growth, photosynthesis, tissue inorganic osmolyte contents, leaf osmotic adjustment capacity and differentially accumulated sugars and amino acids in the stems and leaves. They combined metabolome and transcriptome analyses to explore the role of osmolytes in sweet sorghum’s response to water scarcity, shedding light on key biosynthetic pathways and genes involved in the production of sugars and amino acids that enhance osmotic stress tolerance. Specifically, genes involved in biosynthesis pathways of sucrose (such as SUS1, SUS2, etc.), trehalose (including TPS6), raffinose (such as RAFS2 and GOLS2, etc.), proline (such as P5CS2 and P5CR), leucine and valine (including BCAT2), and arginine (such as ASS and ASL), were upregulated after 20% PEG treatments of 6 and 48 hours. The upregulation of genes related to the biosynthesis of these compounds further supports sweet sorghum’s adaptation to water scarcity and potential industrial applications.
SorghumBase examples:
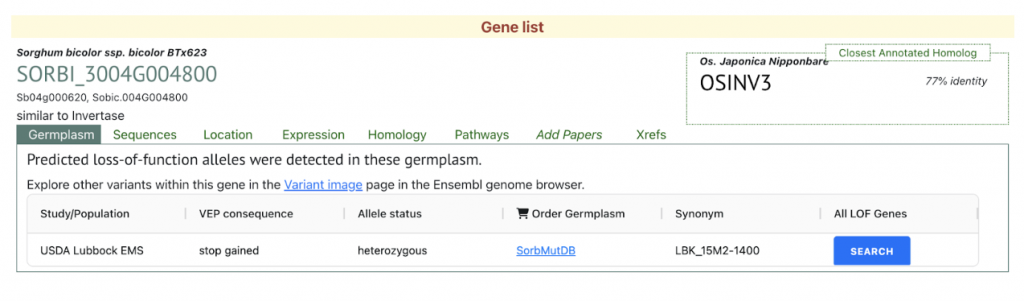
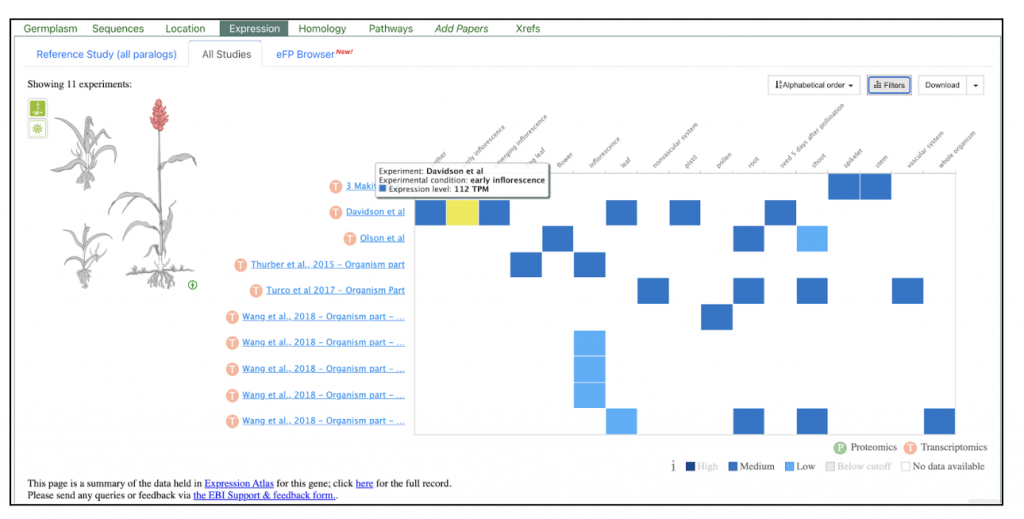
The expression change of DEGs related to sugar metabolism in stems after 20% PEG treatment for 6 hr. Identifies four differentially expressed genes that includes SORBI_3002G188500; SORBI_3002G194700 which are involved in trehalose biosynthesis; SORBI_3004G004800 is involved in sucrose metabolism and SORBI_3001G508800 is involved in starch degradation.
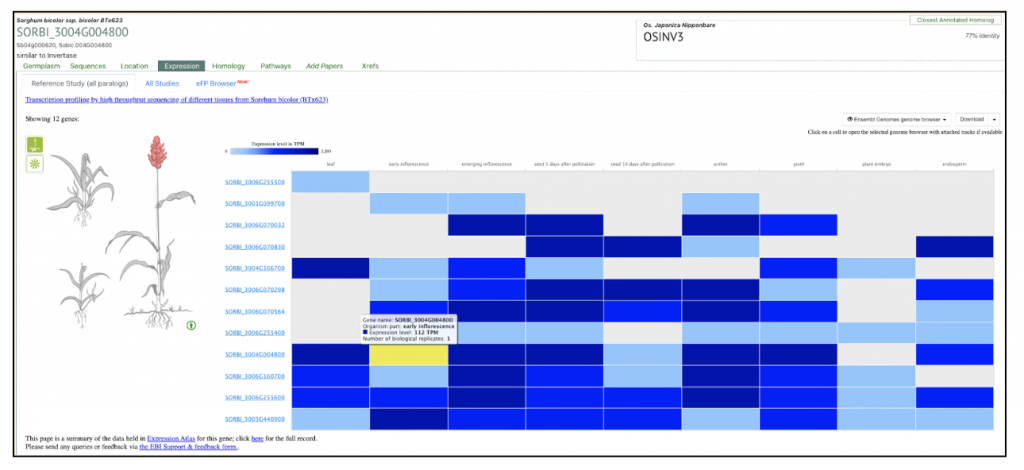
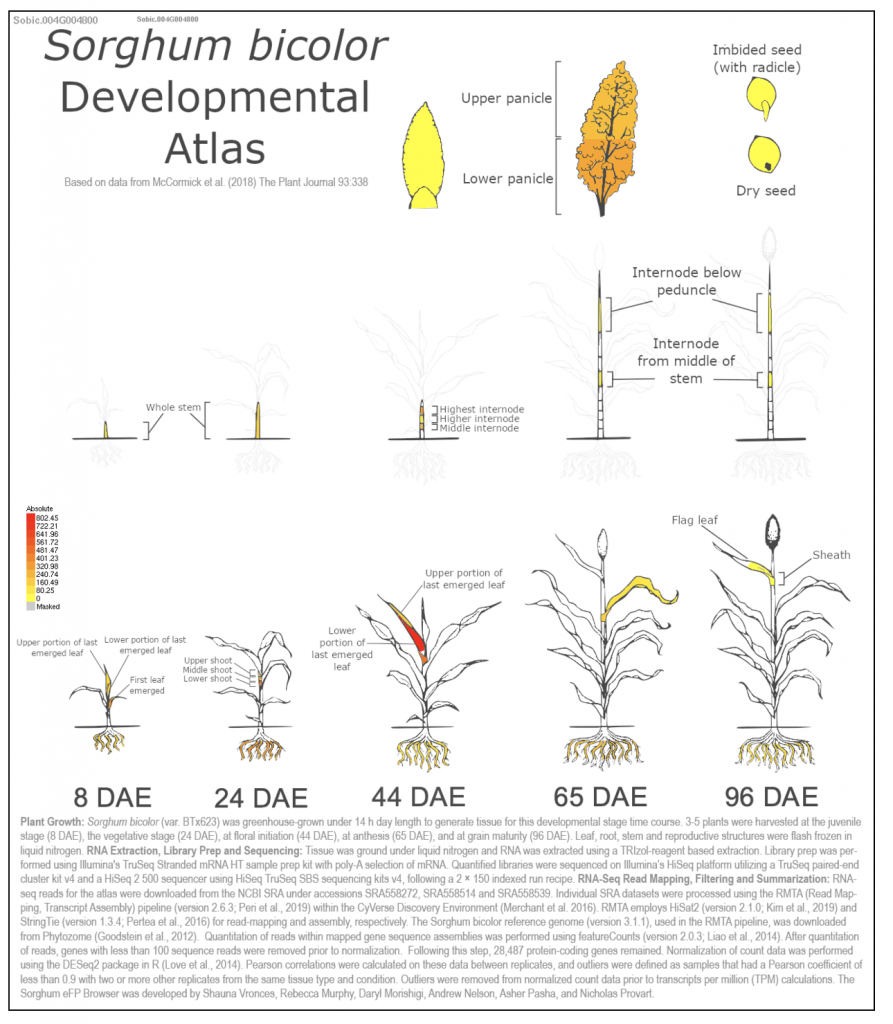
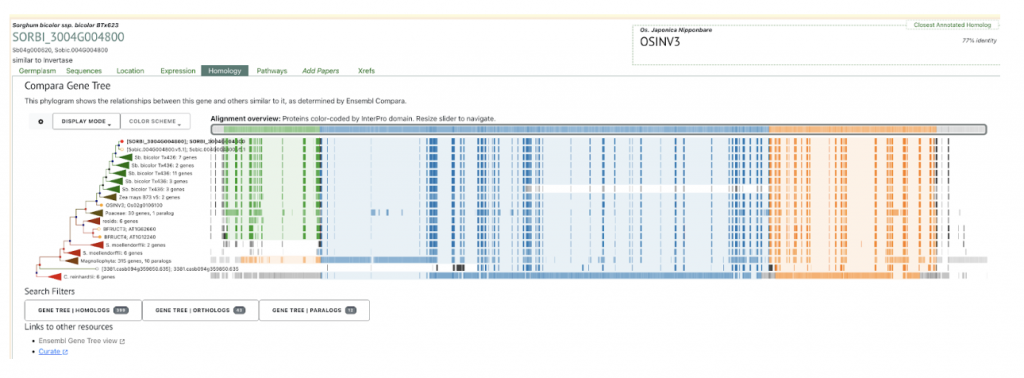
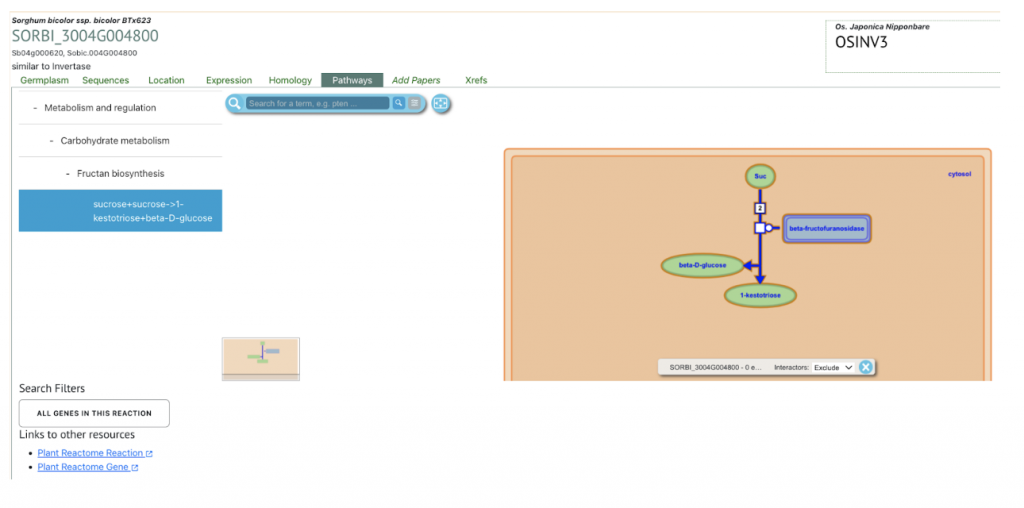
We selected the gene SORBI_3004G004800 (similar to invertase), involved in sucrose metabolism, from this published study as a candidate gene to explore SorghumBase (https://sorghumbase.org/).
Cui YN, Yan SJ, Zhang YN, Wang R, Song LL, Ma Y, Guo H, Yang PZ. Physiological, Metabolome and Gene Expression Analyses Reveal the Accumulation and Biosynthesis Pathways of Soluble Sugars and Amino Acids in Sweet Sorghum under Osmotic Stresses. Int J Mol Sci. 2024 Aug 16;25(16):8942. PMID: 39201628. doi: 10.3390/ijms25168942. Read more