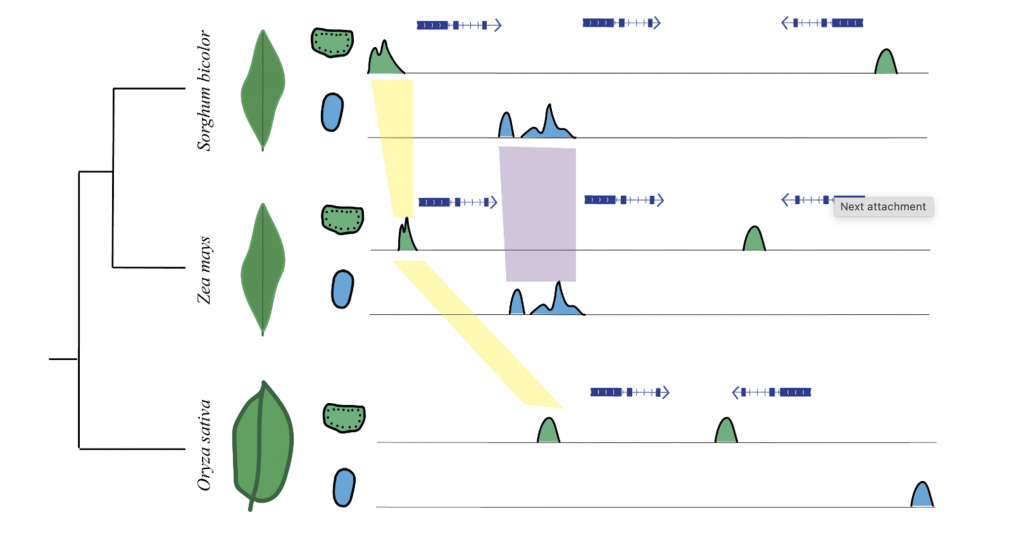
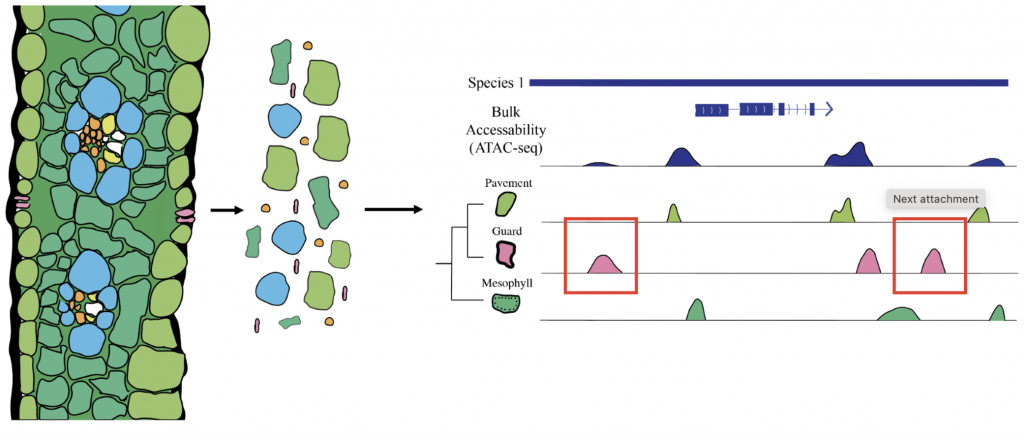
The efficiency of the C3 photosynthetic pathway decreases in hot and arid conditions; C4 photosynthesis has evolved to overcome these challenges by compartmentalizing photosynthetic reactions between mesophyll (MS) and bundle sheath (BS) cells, enhancing CO2 concentration around RuBisCO. This pathway has evolved independently multiple times, suggesting the potential for engineering C3 plants to adopt C4 mechanisms for increased heat tolerance. A key factor in the evolution of C4 photosynthesis is the role of cis-regulatory elements (CREs), which regulate the cell-type-specific expression of critical C4 genes. Previous studies have shown that CREs are crucial in driving C4 gene expression, and recent advances in genomic techniques, such as single-cell ATAC-seq, have allowed the identification of CREs associated with C4 loci in different grass species.
Researchers from University of Georgia, Shanghai Jiao Tong University and The Chinese University of Hong Kong explored the evolution of cis-regulatory elements associated with C4 photosynthesis using single-cell ATAC-seq data. By analyzing chromatin accessibility in cell-type-specific regions from four C4 species and one C3 outgroup, the researchers identified key accessible chromatin regions (ACRs) and transcription factor (TF) motifs associated with C4 enzymes. The findings suggest a mix of conserved and novel ACRs surrounding C4 genes, indicating ongoing regulatory evolution. Cell-type-specific ACRs were particularly enriched around core and subtype-specific C4 enzymes, with some falling outside of the core promoter region. The study also revealed de novo TF binding motifs around C4 loci, showing rapid evolution and highlighting the complex relationship between motif conservation and gene regulation in C4 photosynthesis.
The analysis provides insights into how CREs evolve to regulate the C4 pathway, with evidence suggesting both co-option of pre-existing regulatory elements and the emergence of new ones. These processes were exemplified in subtype-specific C4 enzymes like NAD-MEs in Panicum miliaceum and PEPCK in Urochloa fusca, which showed novel ACRs. The study also pointed out challenges in identifying conserved and novel ACRs due to genetic distances between species and highlighted the need for additional comparisons with more closely related C3 species. Future work, including transgenic experiments and genome editing, will be crucial in pinpointing specific CREs critical for cell-type-specific expression, aiding in the broader understanding of gene regulation in C4 photosynthesis evolution.
The analysis found in this manuscript stands as the first of its kind exploration into single cell chromatin accessibility in a diverse set of species. We hope this manuscript stimulates interest about the rapid rate of cis regulatory evolution in plants and how it can relate to key traits of interest. – Mendieta
SorghumBase examples:
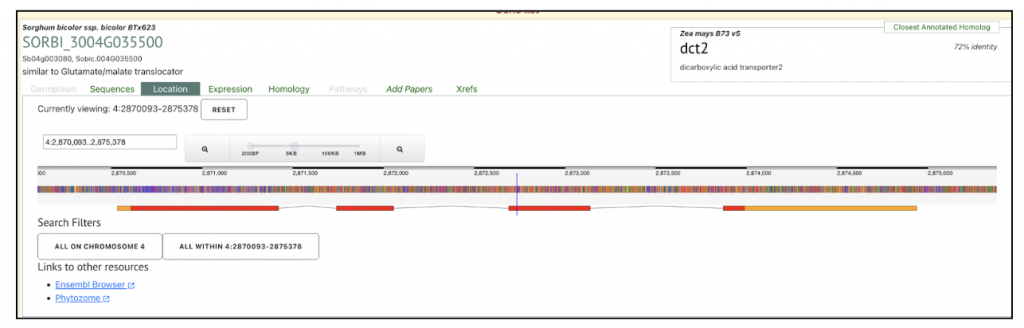
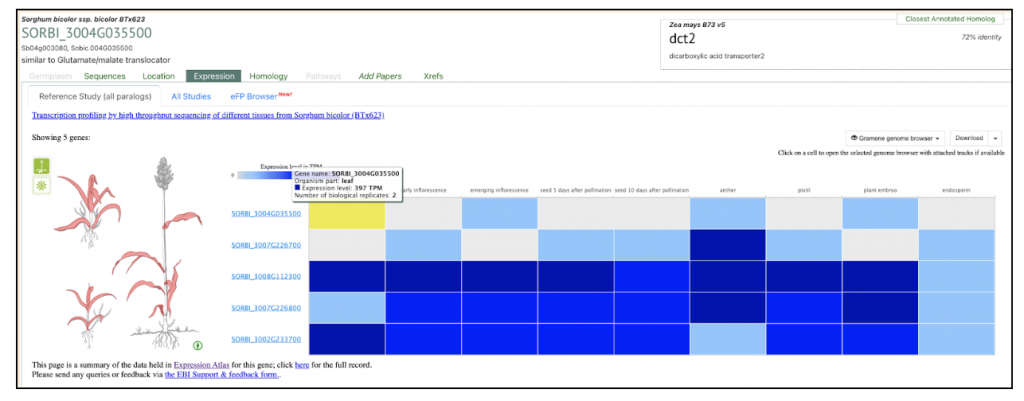
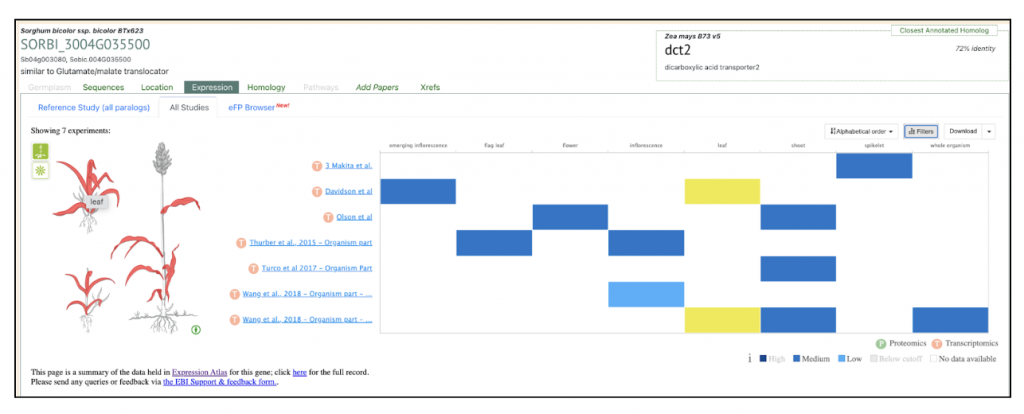
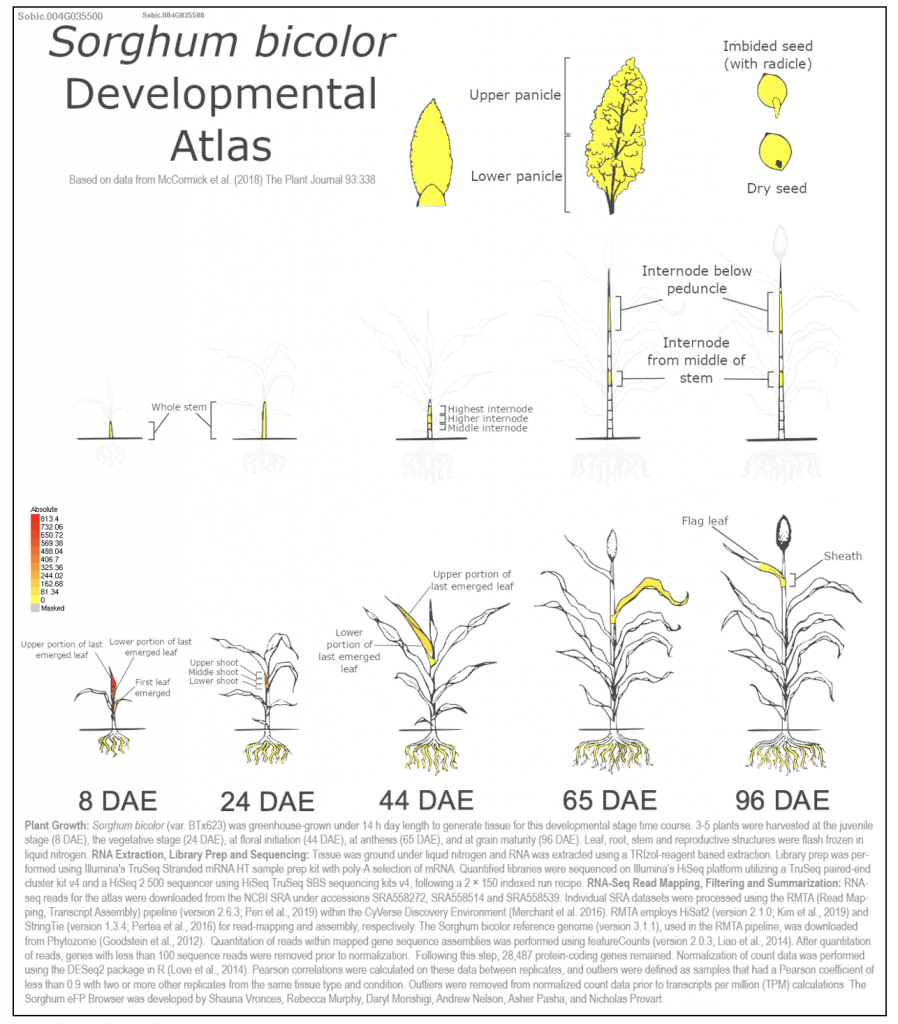
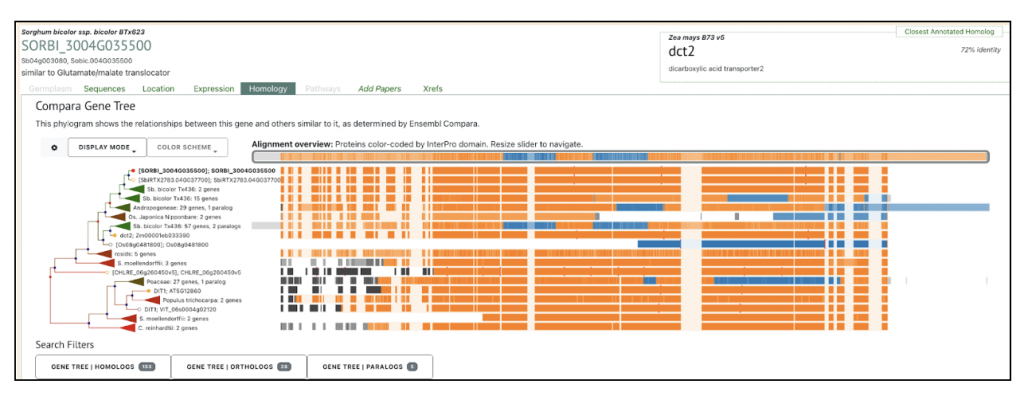
Zea mays dct1 Zm00001eb101270 has two sorghum orthologs SORBI_3004G035500 and SORBI_3002G233700. The paper says “The malate transporters DICARBOXYLIC ACID TRANSPORTER1/2 (DIT1/2) also demonstrated their expected cell-type-specific bias with DIT1 being MS specific and DIT2 being BS specific in both species (Fig. 3 B and C). However, upon further inspection of the phylogenies of the DITs in S. bicolor, we noticed a pattern where the most BS-biased copy, SbDIT4 (Sobic.004G035500), was phylogenetically more closely related to the ZmDIT1, something previously reported (33, 36). These results indicate that over evolutionary time, even members of the same C4 photosynthetic subtype, which likely share a C4 ancestor, can use different paralogous loci to achieve cell-type-specific expression. This highlights that C4 evolution is an ongoing process.”
We selected the gene SbDIT4 (Sobic.004G035500) (Sb04g003080, Sobic.004G035500 similar to Glutamate/malate translocator) from this published study as a candidate gene to explore SorghumBase (https://sorghumbase.org/).

Mendieta JP, Tu X, Jiang D, Yan H, Zhang X, Marand AP, Zhong S, Schmitz RJ. Investigating the cis-regulatory basis of C3 and C4 photosynthesis in grasses at single-cell resolution. Proc Natl Acad Sci U S A. 2024 Oct;121(40):e2402781121. PMID: 39312655. doi: 10.1073/pnas.2402781121. Read more
Related Project Websites:
- Schmitz Lab at University of Georgia: https://schmitzlab.uga.edu/