The evolution of C4 photosynthesis involved the co-option of ancestral transcriptional networks and cis-regulatory elements, enhancing efficiency in carbon fixation and offering insights for engineering C4 traits in C3 crops.
Unraveling the regulatory architecture behind C4 photosynthesis reveals how evolution repurposed ancestral genetic networks to overcome cellular constraints, offering a possible blueprint for engineering photosynthetic traits into staple C3 crops and helping boost global food resilience under climate stress. – Swift
The evolution of C4 photosynthesis represents a significant adaptation in plants, enhancing efficiency in carbon fixation, water use, and nitrogen utilization. While most plants use the ancestral C3 pathway, where RuBisCO fixes CO2 in mesophyll cells, this process is prone to inefficiency due to RuBisCO’s inability to fully discriminate between CO2 and O2, leading to energy-intensive photorespiration. C4 plants, such as maize and sorghum, evolved a compartmentalized mechanism that minimizes photorespiration by concentrating CO2 around RuBisCO in bundle-sheath cells. This reconfiguration, driven by the interplay between mesophyll and bundle-sheath cells, boosts photosynthetic performance, especially in hot, arid environments, making C4 plants some of the most productive crops. Central to this adaptation is the integration of transcriptional networks and cis-regulatory elements, which reprogram cell-type-specific gene expression patterns to support the new photosynthetic pathway.
Recent findings from scientists at Salk Institute for Biological Studies and University of Cambridge highlight the role of DOF transcription factors and their associated cis-elements in facilitating C4 evolution. DOF motifs, enriched in genes expressed in the bundle sheath across C3 and C4 species, suggest that ancestral cell-identity networks were co-opted and expanded during C4 evolution. This regulatory mechanism appears conserved across monocotyledons and dicotyledons, supporting a model where evolution repurposed existing genetic frameworks for novel functions. Insights into these molecular underpinnings have profound implications for engineering C4 traits into C3 crops like rice. By harnessing ancient transcriptional networks and rewiring key genes, researchers could overcome current challenges in robustly expressing C4-specific pathways, potentially revolutionizing crop productivity in resource-limited environments.
SorghumBase examples:
We selected the gene SbDOF8 (Sobic.004G284400) (SORBI_3004G284400, Sb04g032040) similar to Dof zinc finger protein) from this published study as a candidate gene to explore SorghumBase (https://sorghumbase.org/).
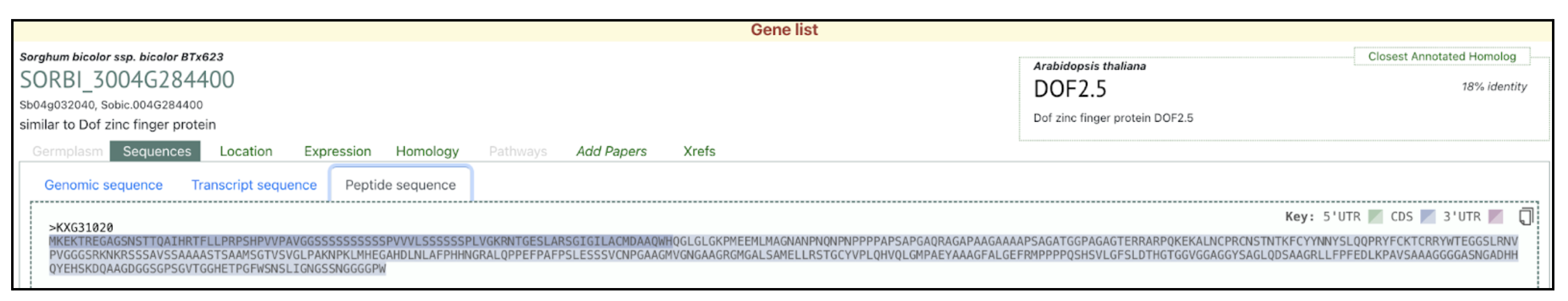
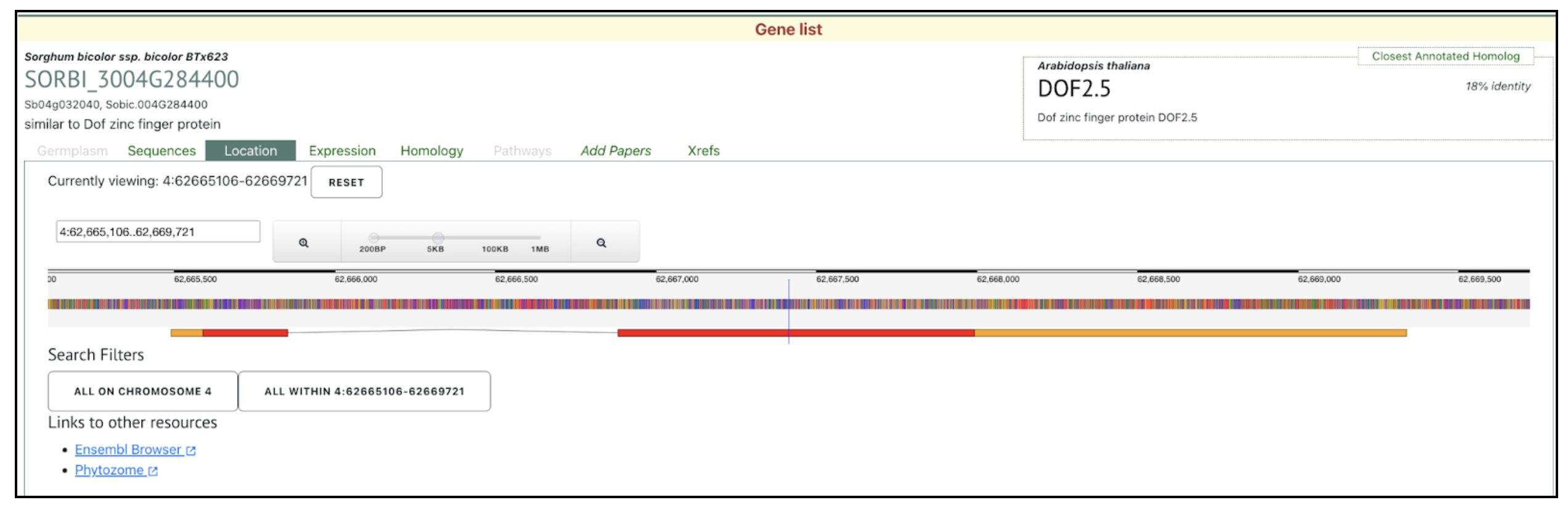

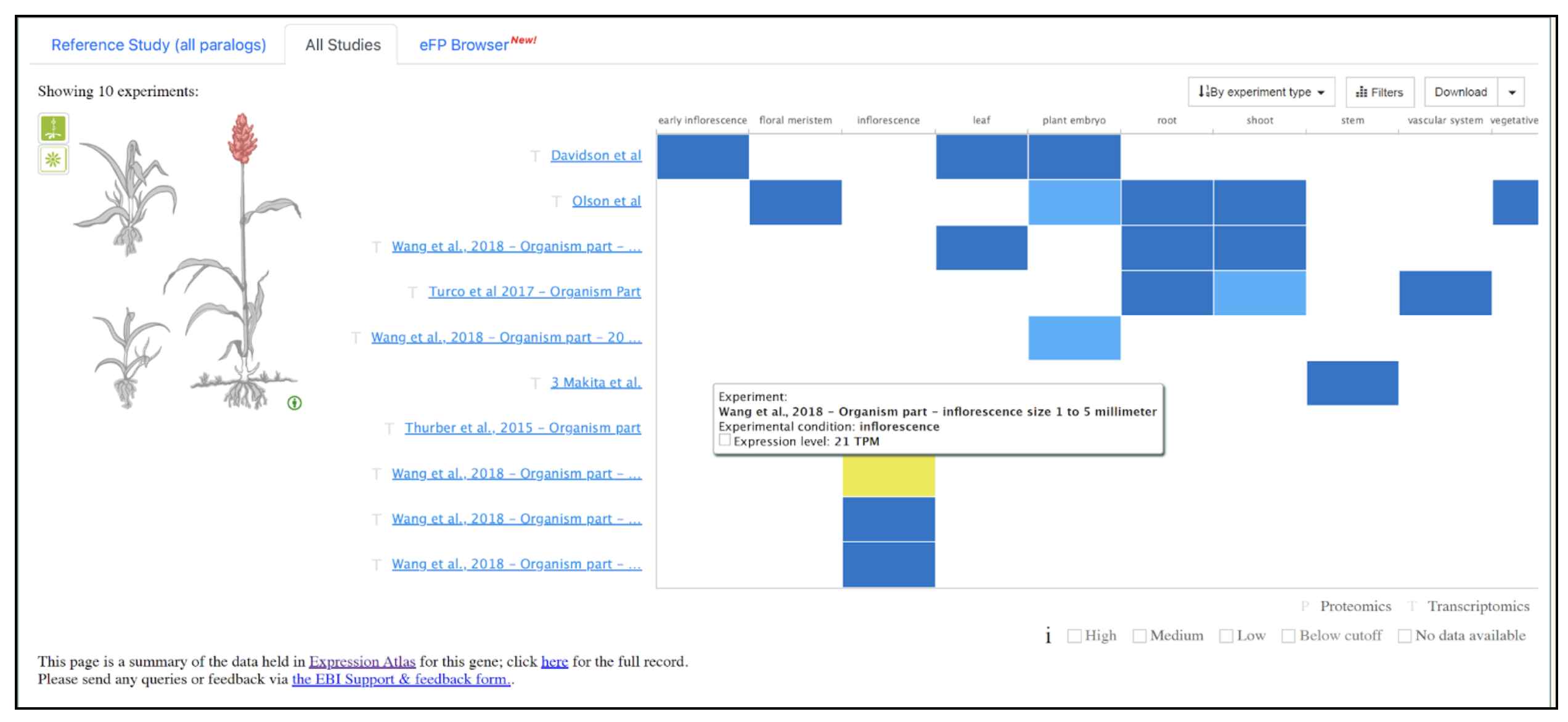

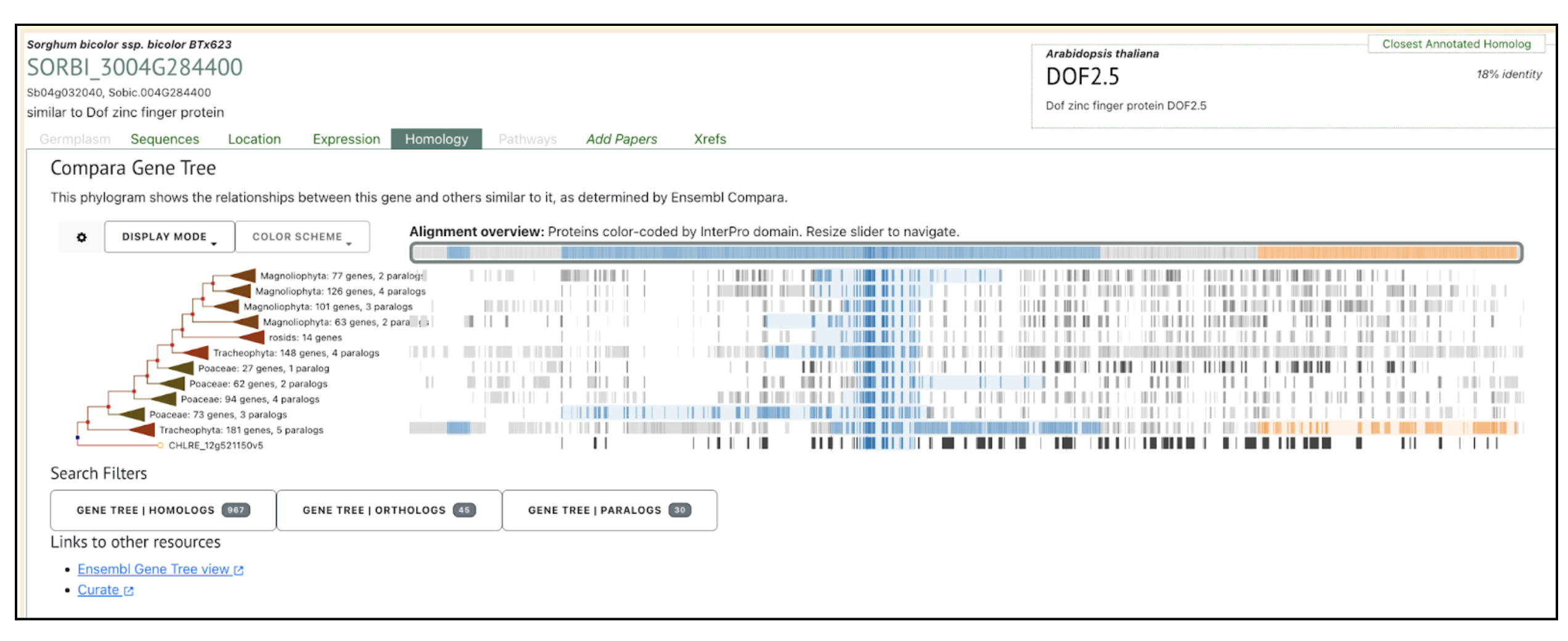
Swift J, Luginbuehl LH, Hua L, Schreier TB, Donald RM, Stanley S, Wang N, Lee TA, Nery JR, Ecker JR, Hibberd JM. Exaptation of ancestral cell-identity networks enables C4 photosynthesis. Nature. 2024 Dec;636(8041):143-150. PMID: 39567684. doi: 10.1038/s41586-024-08204-3. Read more
Related Project Websites:
- Dr. Hibberd’s Molecular Physiology group at the University of Cambridge: https://www.plantsci.cam.ac.uk/research/groups/molecular-physiology
- Ecker lab at Salk Institute for Biological Studies: https://ecker.salk.edu/people/