H3K36me3 plays a crucial role in plant development by activating gene expression and antagonizing repressive chromatin marks like H3K27me3 and H2A.Z, thereby shaping developmental and tissue-specific transcriptional programs.
Keywords: H2A.Z, H3K27me3, H3K36me3, Photosynthesis genes, Sorghum bicolor
Gene regulation via H3K36me3 is vital for plant development, as demonstrated by the severe pleiotropic effects caused by the K36M mutation in Arabidopsis. This histone modification is catalyzed by SET domain group (SDG) proteins, with at least three key enzymes identified in both Arabidopsis (SDG7, SDG8, SDG24, SDG26) and rice (SDG708, SDG724, SDG725). These enzymes regulate numerous developmental processes, including germination, flowering, and reproductive organ development. H3K36me3 deposition is conserved across plant species, facilitating gene activation through interactions with transcription factors like BES1 in Arabidopsis and OsSUF4 in rice. However, a full understanding of the transcription factors mediating H3K36me3 targeting remains incomplete. Researchers from Three Gorges University and Huazhong Agricultural University sought to uncover histone methylation patterns and identify sorghum methyltransferases. One key finding is that H3K36me3 appears to antagonize other chromatin marks such as H3K27me3 and H2A.Z, suggesting a regulatory balance that shapes gene expression landscapes during development.
In contrast, H3K27me3—typically associated with gene repression—exhibits both short and long enrichment regions in plant genomes. While generally limited to single genes in Arabidopsis, extended H3K27me3 domains spanning multiple genes are more frequent in sorghum, where four large H3K27me3 islands have been identified. The mechanisms underlying this spreading remain unclear but may involve Polycomb response elements or nucleation sites, similar to those in animals. Importantly, H3K36me3 and H3K27me3 show mutually exclusive distribution, with H3K36me3 forming gaps within long H3K27me3 regions, yet not blocking H3K27me3 spread. Furthermore, H3K36me3 enrichment correlates with diminished H2A.Z presence, hinting at a role in chromatin remodeling and possibly in recruiting DNA methylation machinery. These antagonistic interactions highlight a complex interplay between chromatin modifications, modulating gene activity across tissues and developmental stages.
SorghumBase examples:
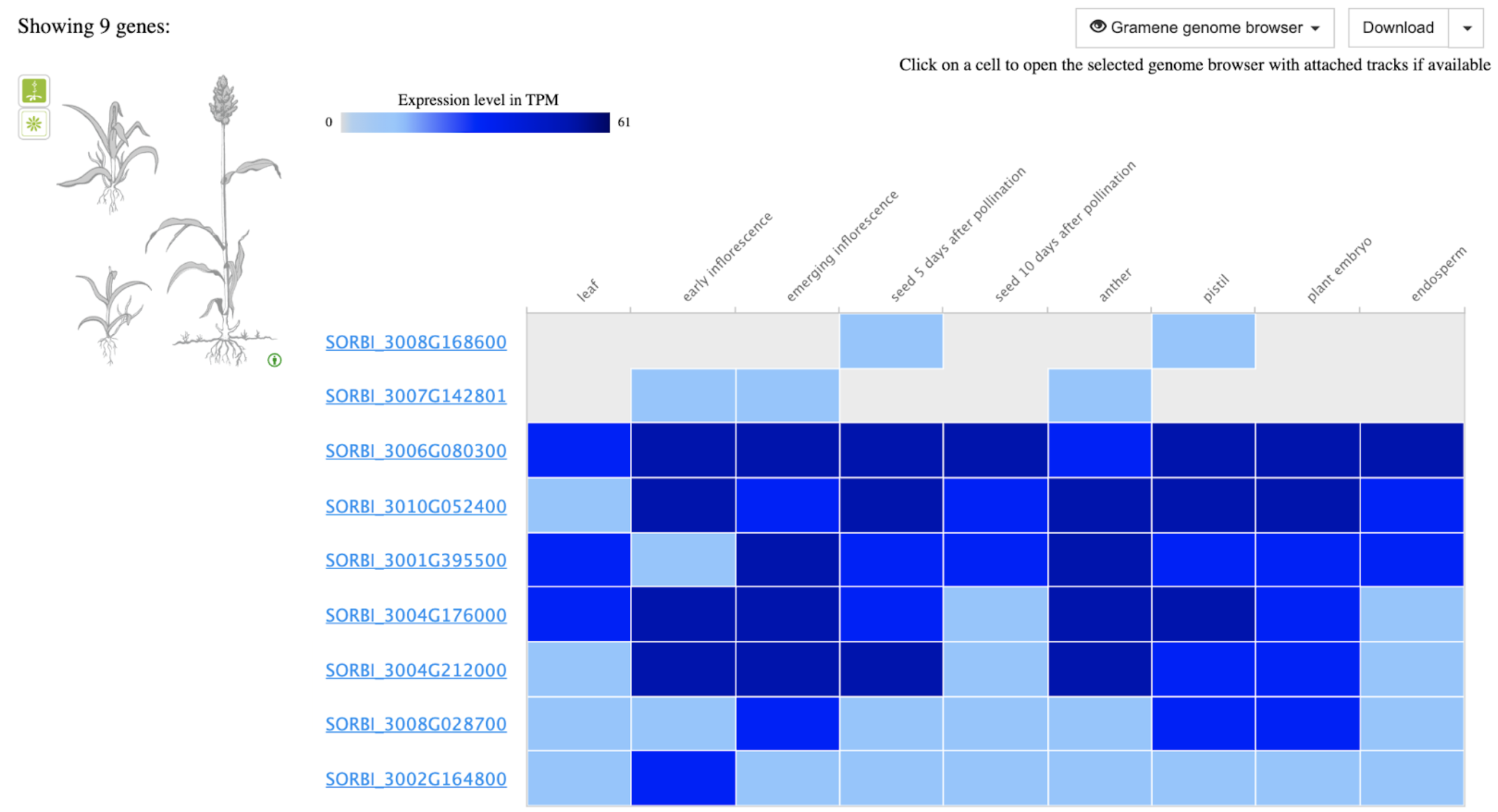
Methyltransferases from this study are part of a larger gene family with 9 paralogs.
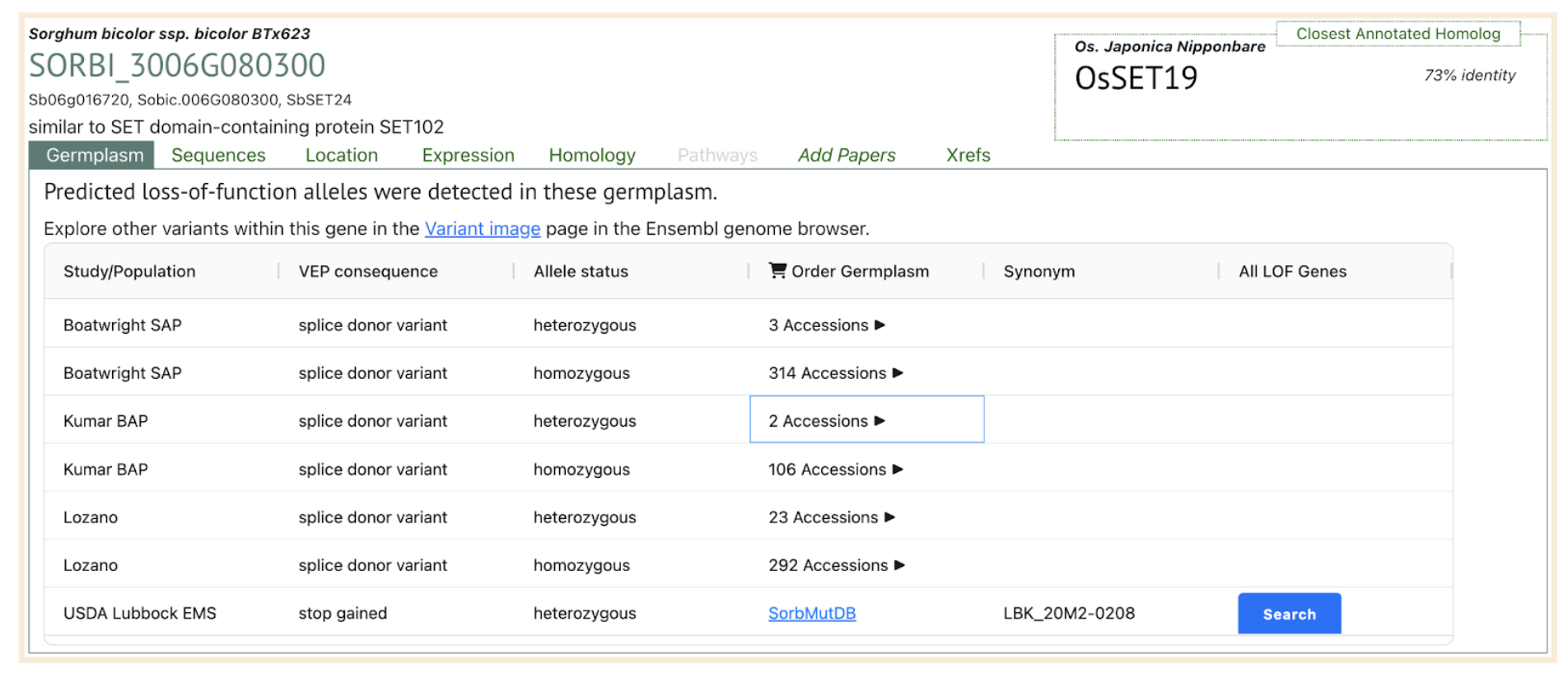
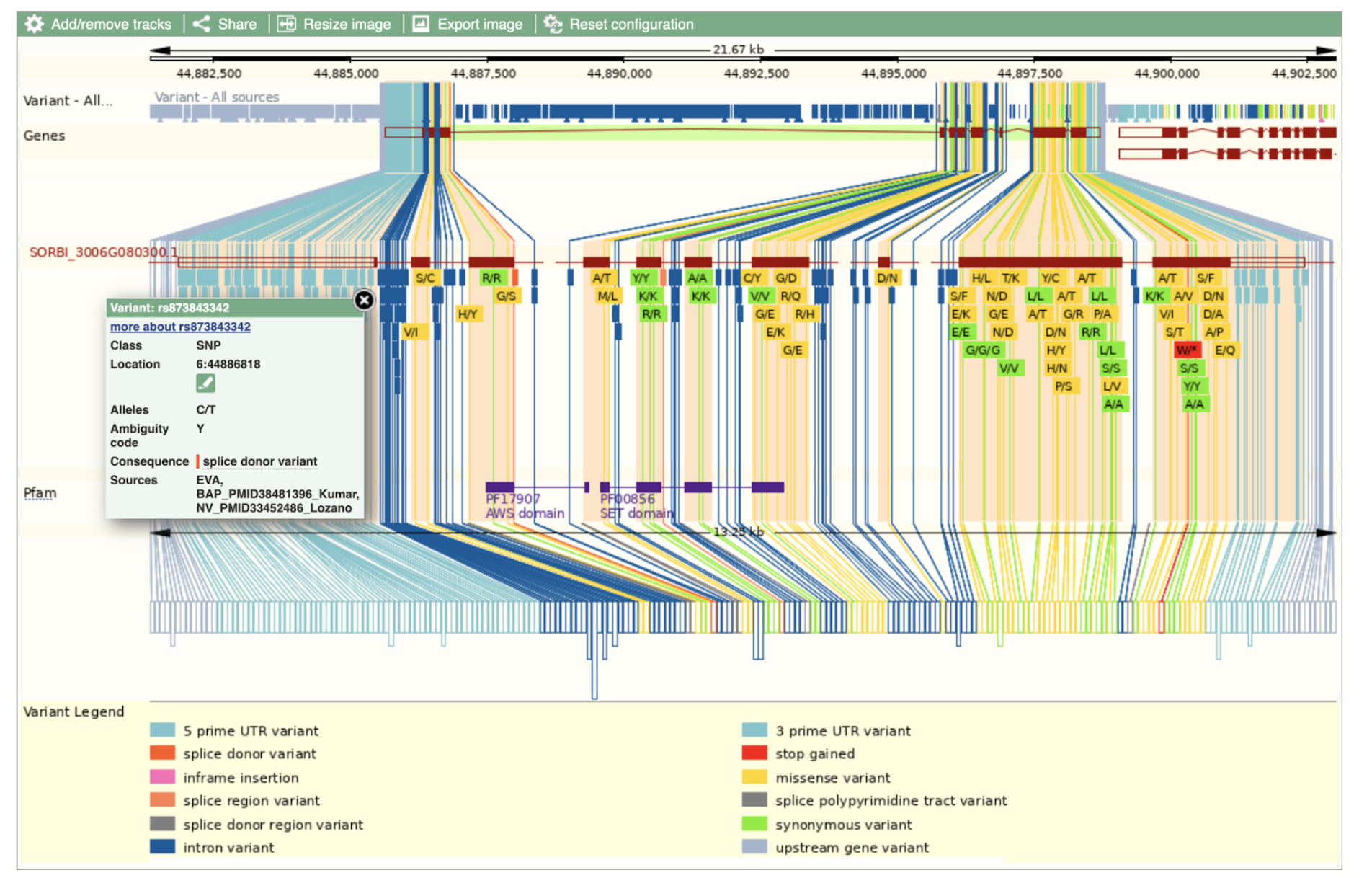
SbASHH1: SORBI_3006G080300
SbASHH2: SORBI_3004G176000
SbASHH3: SORBI_3002G164800

Reference:
Yuan F, He C, Gong X, Zeng G, Qin X, Deng Z, Shen X, Hu Y. H3K36me3 regulates subsets of photosynthesis genes in Sorghum bicolor potentially by counteracting H3K27me3 or H2A.Z. Plant Physiol Biochem. 2025 Mar 24;223:109831. PMID: 40157144. doi: 10.1016/j.plaphy.2025.109831. Read more