Ao et al. identified a BPM domain-containing gene on chromosome 10 as a key regulator of panicle exsertion in sorghum, linking impaired BR signaling and reduced parenchyma cell elongation to a sheathed panicle phenotype, with implications for breeding varieties suited for mechanized harvesting.
Sorghum (Sorghum bicolor), the fifth most cultivated cereal globally, is a C4 plant with superior photosynthetic efficiency, stress tolerance, and adaptability to arid environments. Panicle exsertion (PE)—the elongation of the uppermost internode—is critical for grain yield, pollination efficiency, and mechanized harvesting. In this study, scientists from Guizhou University and Guizhou Academy of Agricultural Sciences identified a sheathed panicle (shp-I) mutant with significantly shortened PE from an EMS-mutagenized population of the variety Hongyingzi (HYZ). Compared to wild-type plants, the mutant displayed reduced plant height, grain size, and internode elongation, primarily due to impaired elongation of parenchyma cells. Histological analysis confirmed decreased cell length and width, similar to defects observed in rice PE mutants such as sui2 and OsWRKY78, highlighting the conserved role of parenchyma extensibility in grass internode development.
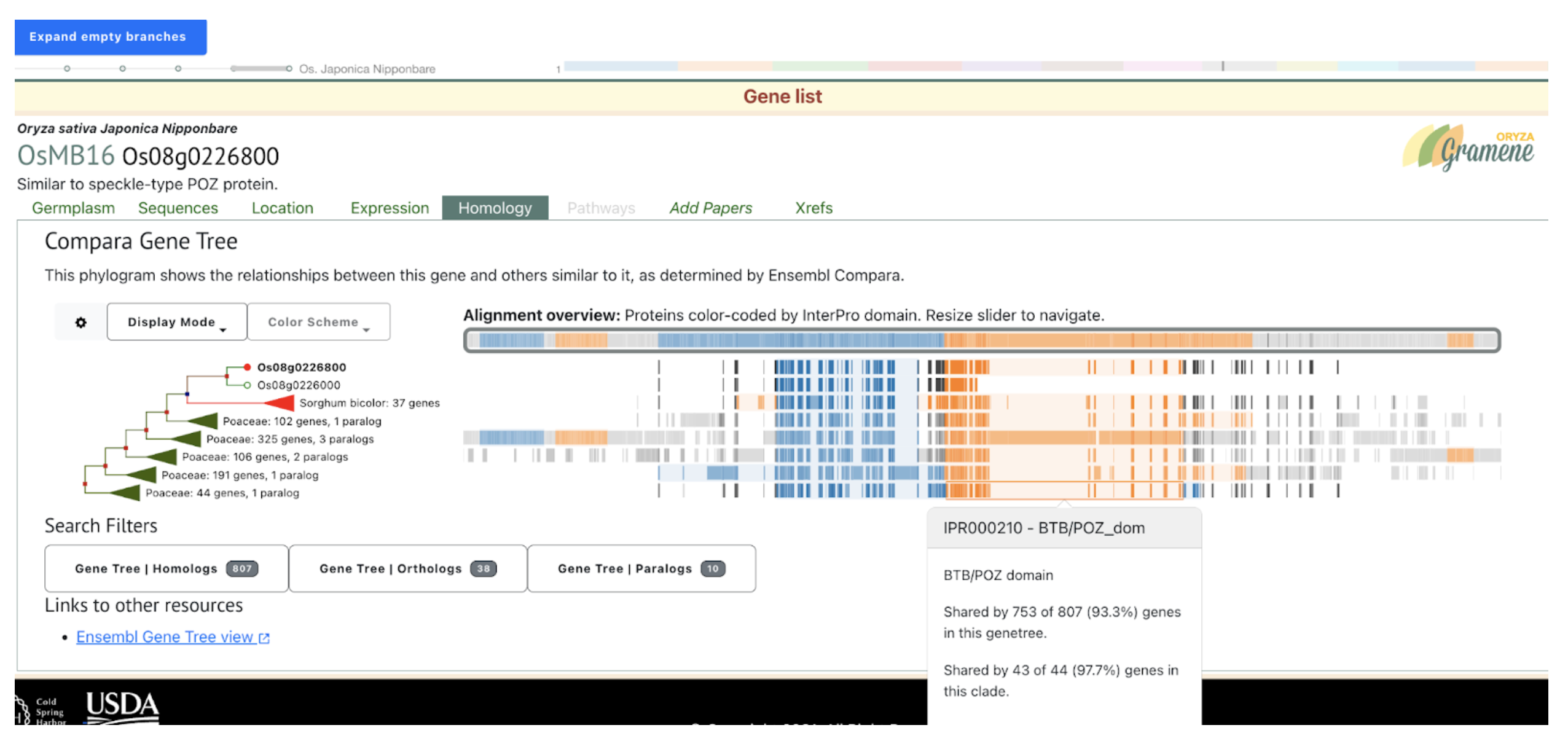
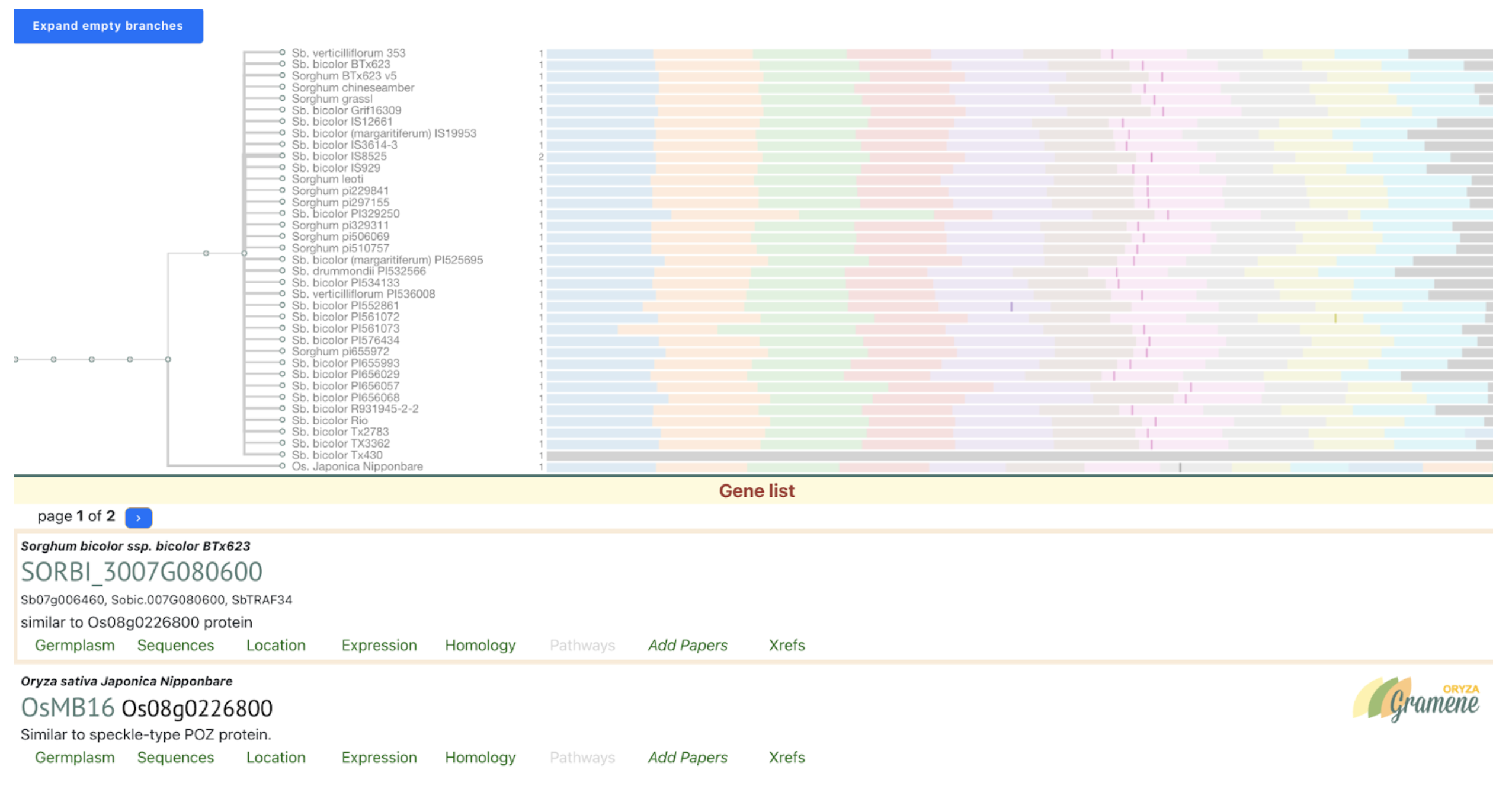
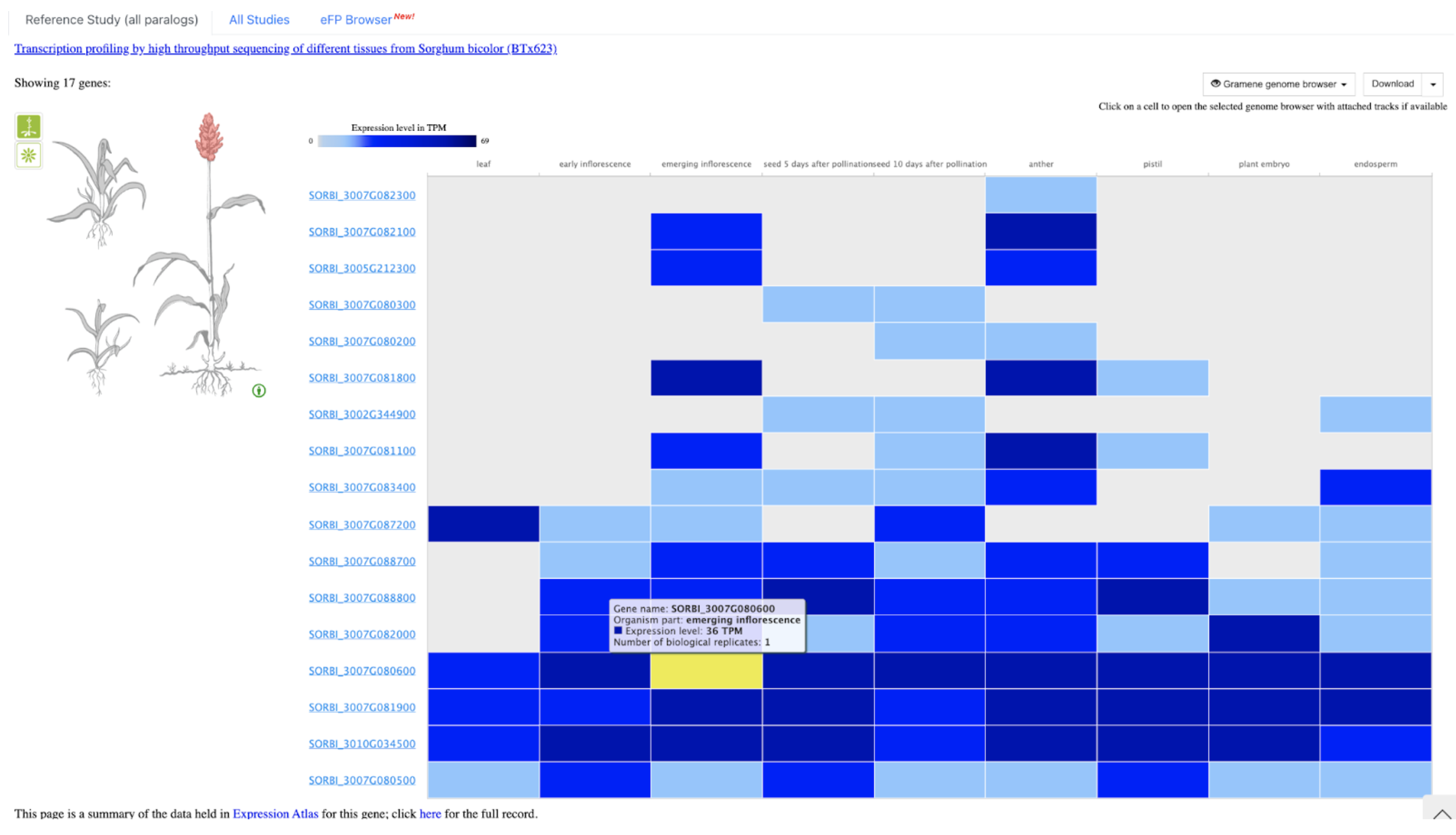
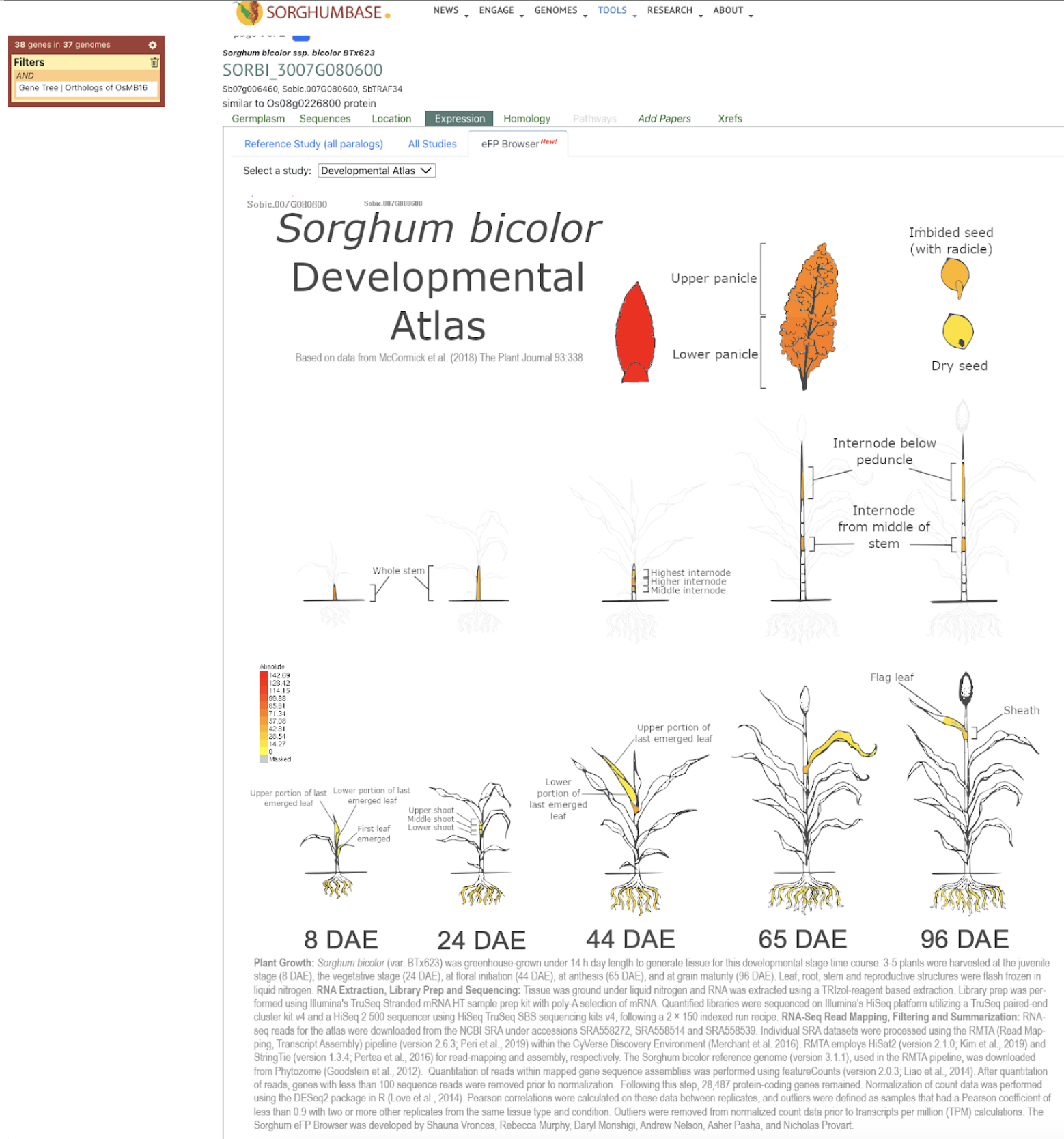
Further analysis revealed that brassinosteroids (BR) and auxin (IAA) play central roles in regulating PE elongation. Although gibberellin (GA3) levels were unchanged in the mutant, BR concentration was elevated, likely due to disrupted BR signaling, triggering feedback upregulation of BR biosynthesis. This BR overaccumulation appears to suppress auxin levels, impairing cell elongation. Genetic mapping localized the causal locus to chromosome 10, overlapping with previously reported QTLs. A candidate gene, SbiHYZ.10G230700, encoding a BPM domain-containing protein—a component of the E3 ubiquitin ligase complex—was identified. Mutations within its MATH domain may impair substrate recognition, disrupting BR signaling and PE development. This discovery provides valuable insights into sorghum internode elongation and offers a molecular target for breeding varieties suited for efficient mechanical harvesting.
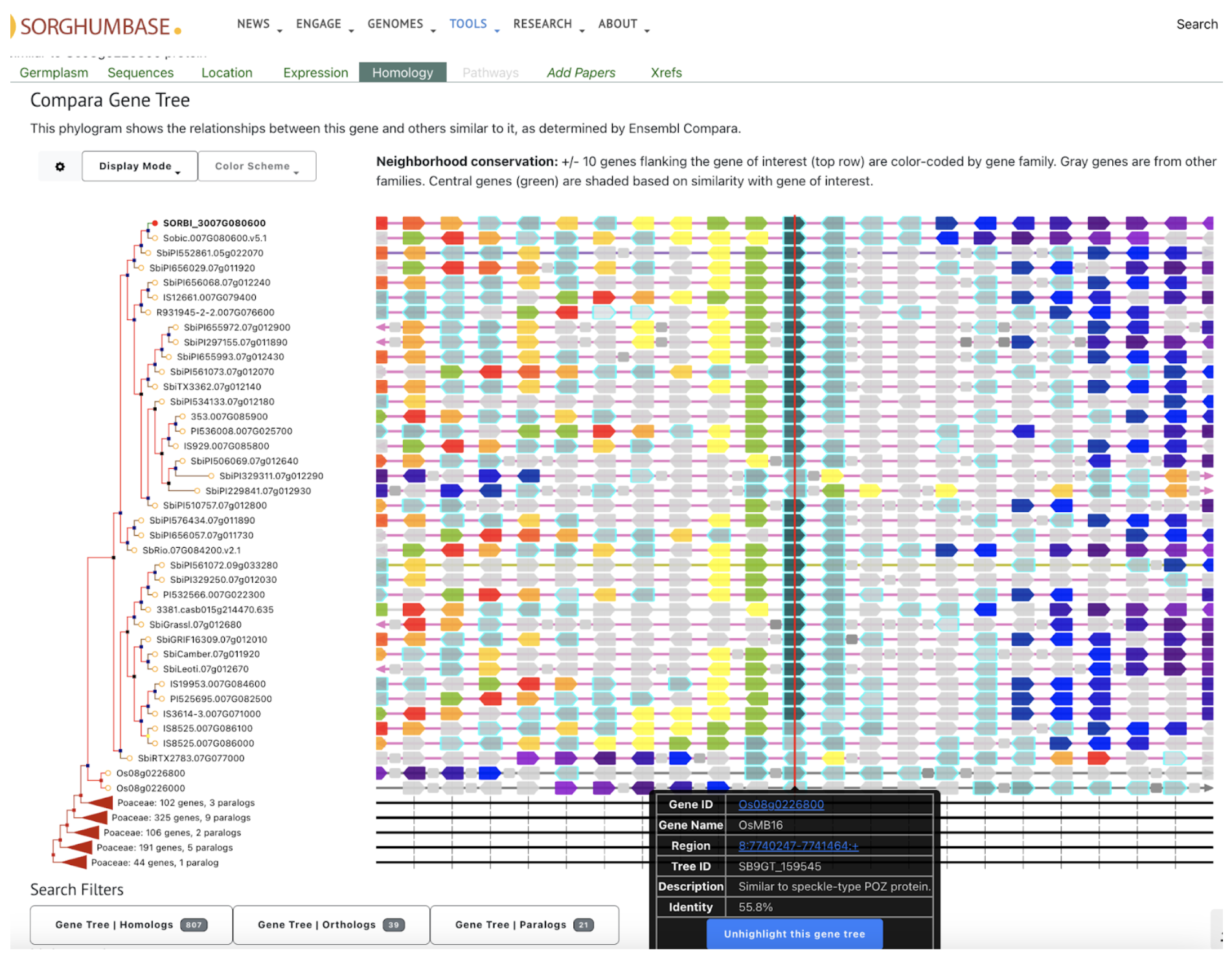
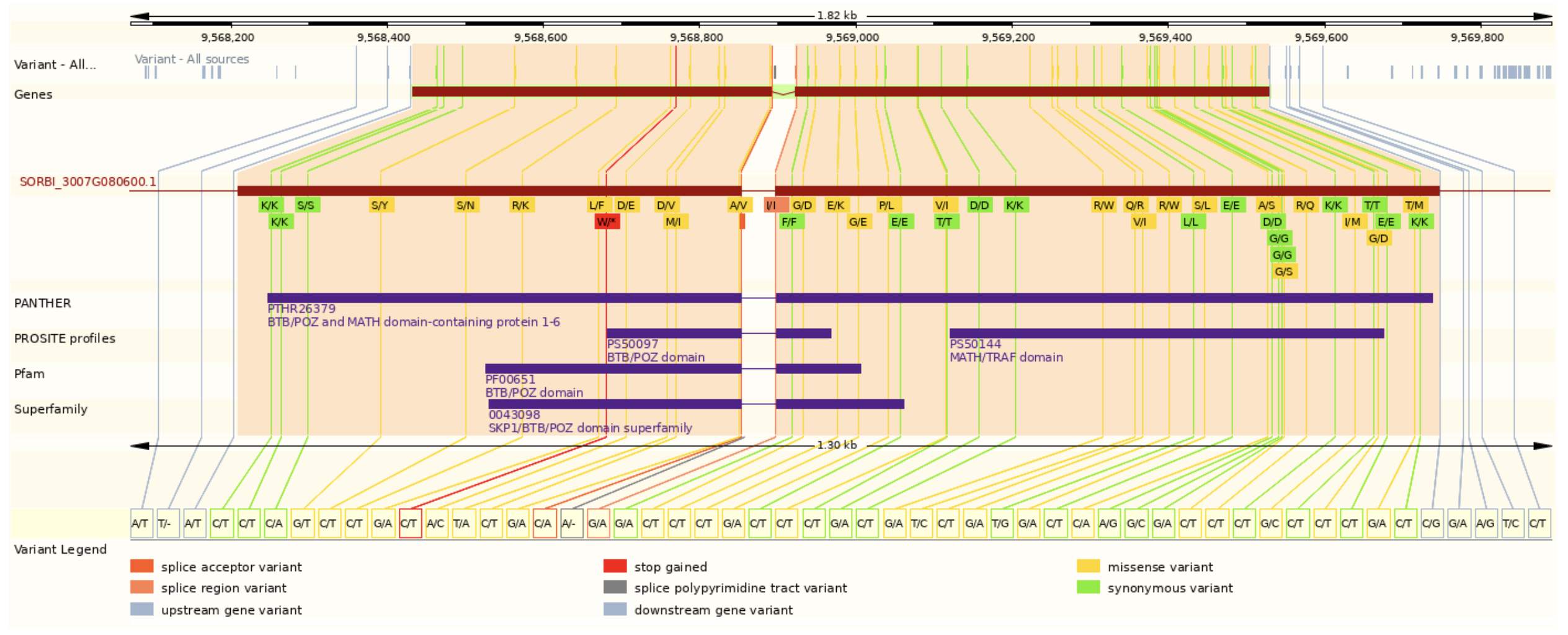
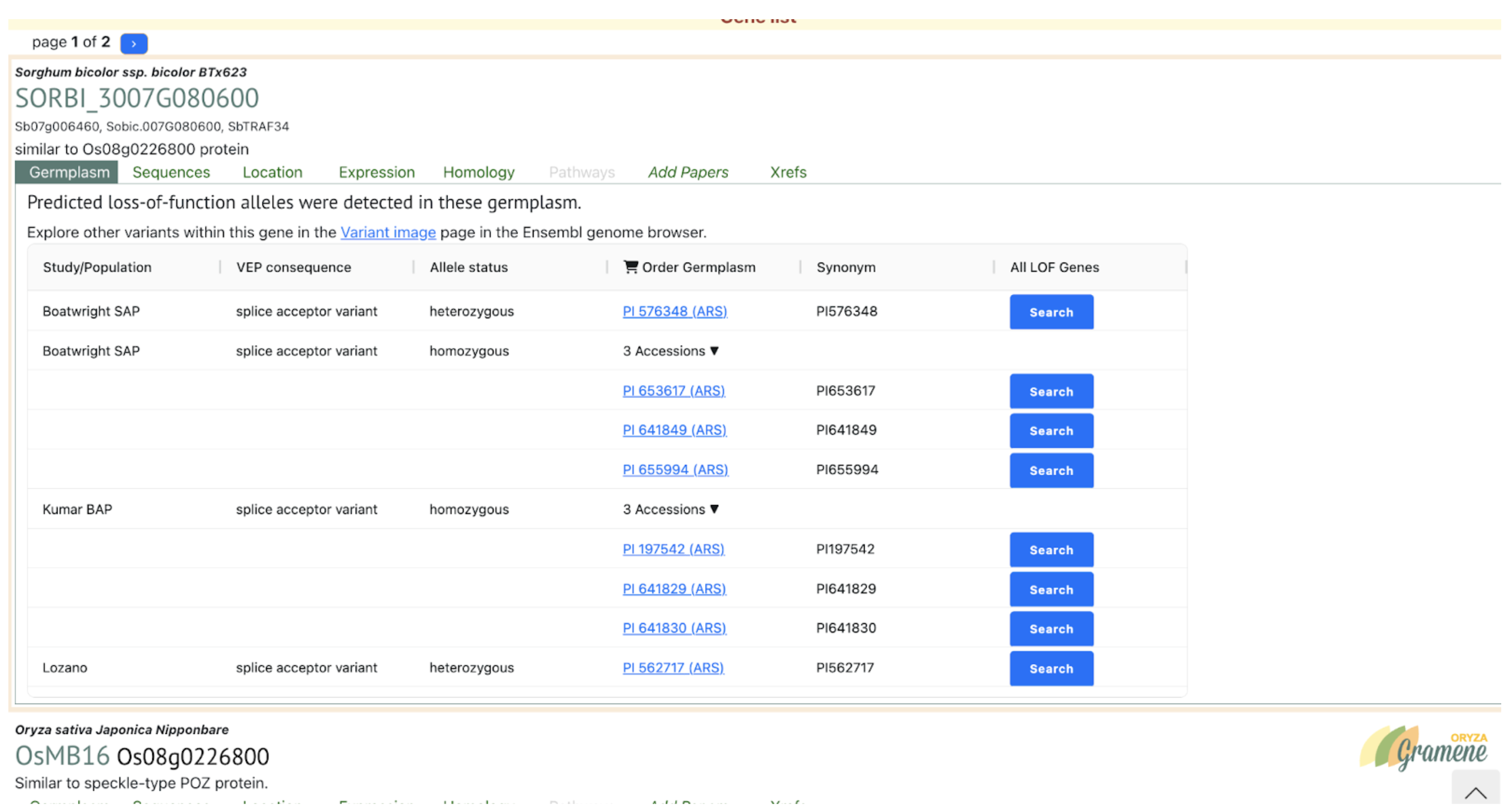
SorghumBase examples:
Reference:
Ao J, Wang R, Li W, Ding Y, Xu J, Cao N, Gao X, Cheng B, Zhao D, Zhang L. Gene mapping and candidate gene analysis of a sorghum sheathed panicle-I mutant. Plant Genome. 2025 Mar;18(1):e70007. PMID: 40032606. doi: 10.1002/tpg2.70007. Read more