Katamreddy et al., revealed that drought-induced hydrogen cyanide (HCN) accumulation in sorghum is regulated by genotype-specific differences in dhurrin biosynthesis, membrane stability, and transcription factor networks, offering targets for developing safer, drought-tolerant forage varieties.
Keywords: HCN potential, WGCNA, dhurrin biosynthesis, drought stress, membrane damage, metabolomics, transcriptomics
Drought stress (DS) in sorghum significantly influences the production of hydrogen cyanide (HCN), a toxic compound derived from the cyanogenic glucoside dhurrin. Dhurrin acts as a defense metabolite and is primarily stored in vacuoles, releasing HCN upon cellular damage. Environmental stresses such as drought induce membrane disruption, allowing dhurrin to contact activating enzymes and release HCN, posing a toxicity risk to livestock. In the present study, researchers from ICRISAT and collaborating institutions demonstrated that drought-sensitive sorghum genotypes exhibit high HCN production due to enhanced expression of key genes in the cyanoamino acid metabolism pathway (e.g., CYP79A1, CYP71E1, UGT85B1), driven by drought-induced membrane damage and stress signaling. In contrast, drought-tolerant genotypes show suppressed expression of these genes, likely through transcriptional repressors (e.g., MYB, Dof), suggesting protective regulatory mechanisms that mitigate dhurrin bioactivation and limit HCN accumulation under drought.
Transcriptomic and metabolomic profiling revealed that drought tolerance in sorghum correlates with the upregulation of genes involved in membrane repair, detoxification (e.g., PER1, EIN3), and stress-responsive metabolic pathways such as amino acid degradation and jasmonic acid synthesis. These pathways maintain cellular integrity and reduce the activation of defense responses like cyanogenesis. Regulatory network analysis highlighted the role of transcription factors (TFs), especially ERFs, in positively regulating HCN biosynthetic genes in sensitive genotypes under drought. This integrative approach underscores that HCN production in sorghum is not solely a direct consequence of drought but is mediated by the severity of cellular damage and genotype-specific responses. The identification of key regulatory genes provides potential targets for molecular breeding, including CRISPR-based strategies, to develop acyanogenic, drought-resilient sorghum lines suitable for safer livestock forage.
SorghumBase examples:
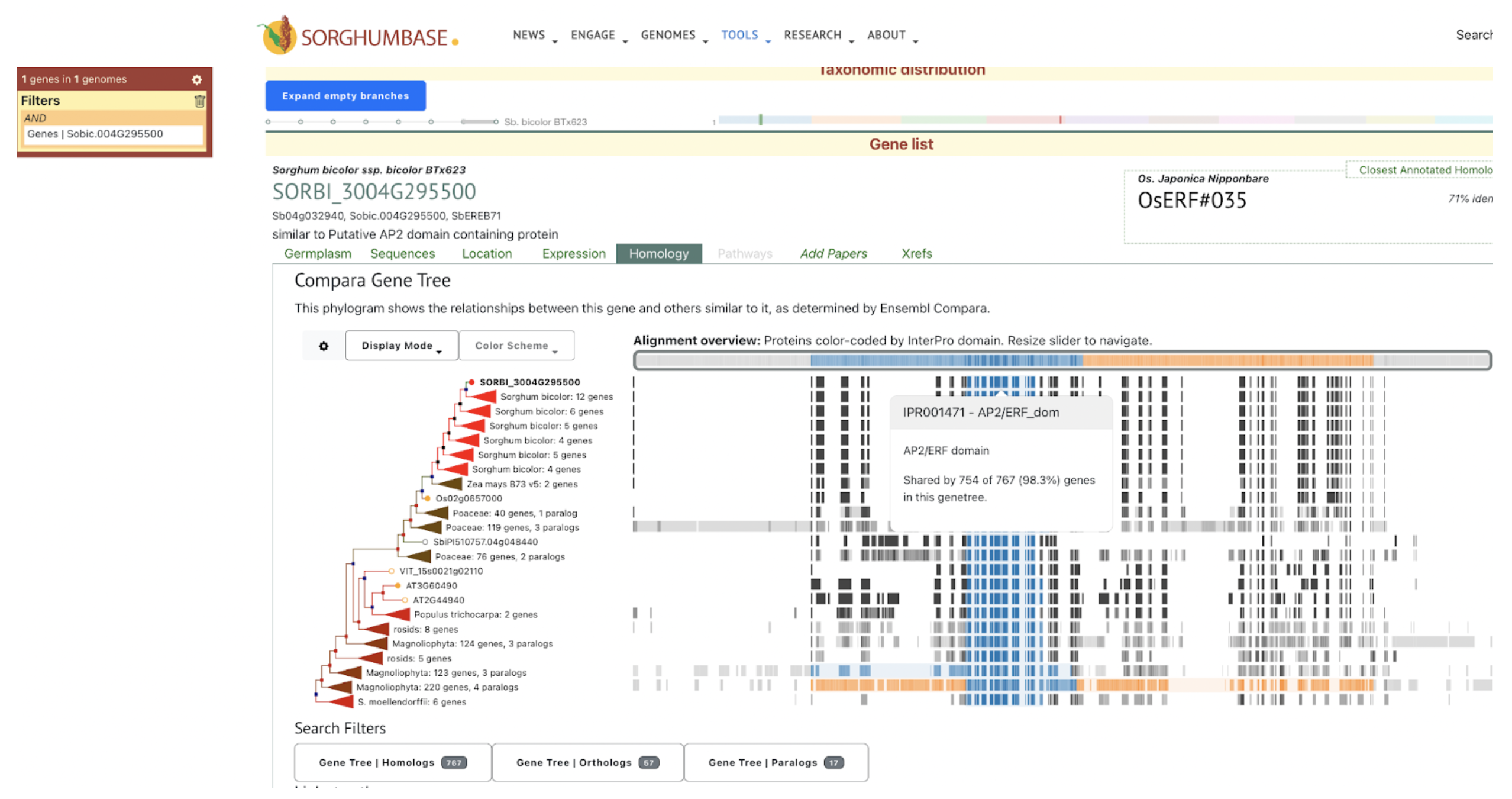
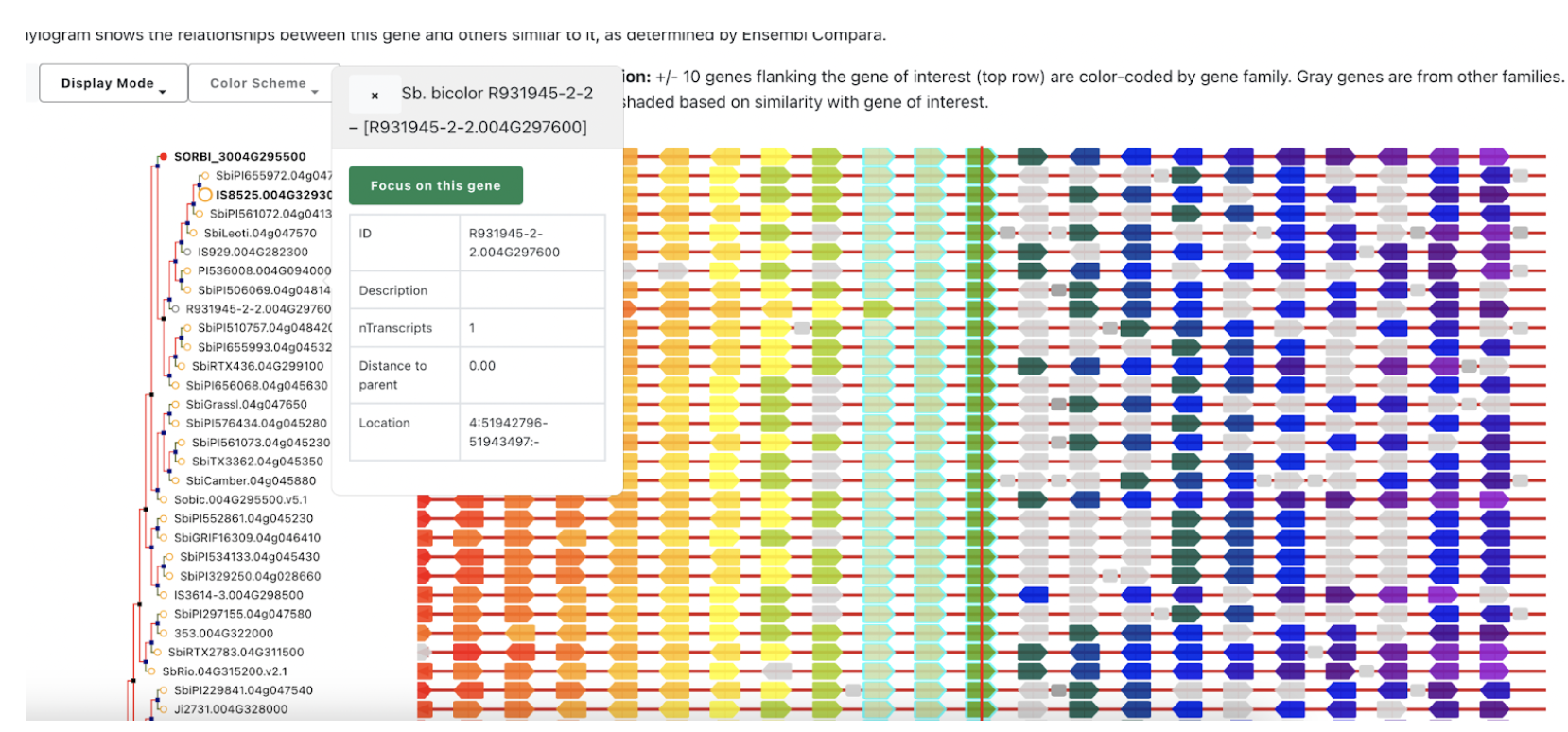
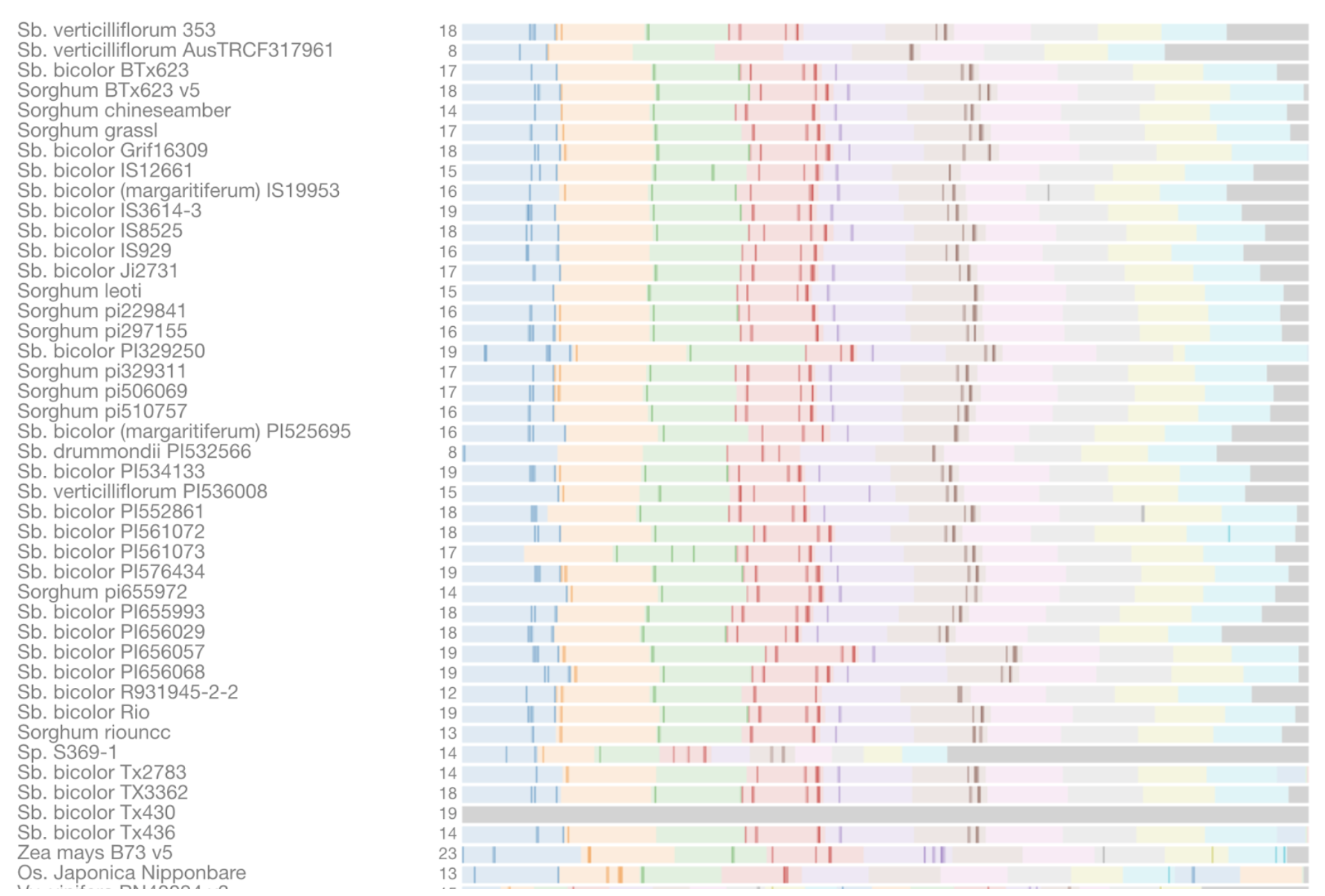
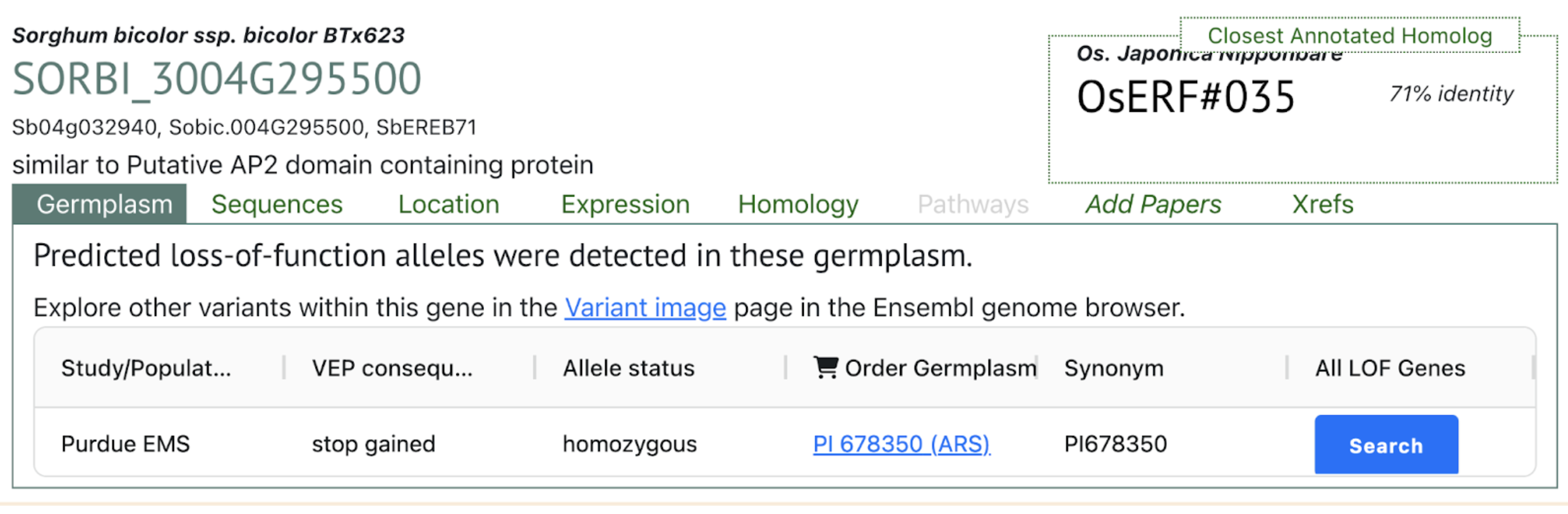
The TF network of HCN‐related genes showed that ERF (Sobic.004G295500) is critical for expressing cyanoamino acid metabolism gene UGT85B1 (Sobic.001G012400) and HCN degradation gene thiosulfate thiotransferase (SEN1‐ Sobic.010G270300) under DS in the ICSR 14001 genotype.
Reference:
Katamreddy SC, Jose J, Reddy BP, Sivasakthi K, Sanivarapu H, Dube N, Gaddameedi A, Kodukula V S V P, Yogendra K, Govindaraj M, Kholova J, Roy Choudhury S, Kishor PBK, Varijakshapanicker P, Mathur PB, Kumar AA, Reddy PS. Drought-Induced Cyanogenesis in Sorghum (Sorghum bicolor L.): Genotypic Variation in Dhurrin Biosynthesis and Stress Response. Plant Cell Environ. PMID: 40214306. doi: 10.1111/pce.15536. Read more