Researchers identified novel genetic regulators of flowering time in temperate-adapted, photoperiod-insensitive sorghum using expanded genome-wide and transcriptome-wide association analyses, revealing key roles for FT, MADS-box, and ageing pathway genes beyond the classical maturity loci.
Keywords: MADS box transcription factors, SBP transcription factors, aging pathway, flowering time, genome‐wide association study, micro-RNAs, photoperiod pathway, sorghum bicolor, transcriptome wide association study
GWAS is a great tool for finding regions of the genome, but particularly in species with elevated linkage disequilibrium, like sorghum, it isn’t great for identifying specific candidate genes. What really excited me about this project was how Harshita Mangal and the team were able to integrate GWAS and TWAS to identify specific candidate genes linked to flowering time variation in temperate sorghum. Since we published the complete set of gene expression and genetic marker data, I’m hoping other people will be able to build on these resources to identify genes controlling variation in other traits in sorghum. – Schnable
By expanding the number of individuals analyzed, we gained deeper insights into the regulation of complex biological processes like flowering time. The high-quality dataset released with this paper will equip scientists to refine and improve lines for an uncertain future. – Torres-Rodriguez
The adaptation of sorghum to temperate environments has largely relied on introgression of photoperiod insensitivity into tropical germplasm, yet significant variation in flowering time remains among temperate-adapted lines. Researchers from University of Nebraska-Lincoln and Alabama A&M University addressed the limitations of prior association mapping efforts—such as small population sizes and overlooked allelic diversity—by leveraging a larger panel of 738 photoperiod-insensitive sorghum conversion lines. Through combined genome-wide (GWAS) and transcriptome-wide (TWAS) association analyses, the study identified multiple candidate loci influencing flowering time that were distinct from the six classical maturity genes. Key findings included strong associations with the FT-family genes SbFT8 and SbFT2, as well as two MADS-box genes (SbMADS7 and SbMADS31) involved in inflorescence meristem identity. Additionally, several components of the aging pathway were implicated, most notably SbSBP19 and SbSBP4, which are potential targets of mir156, a microRNA known to delay flowering in model species. A signal near SBI-MIR172a and its predicted target rap2.7 ortholog further supports the role of conserved age-related regulatory networks in sorghum flowering time.
Beyond known pathways, this work uncovered associations with genes linked to developmental timing, meristem function, and leaf senescence, such as SbNAC32, tornado1, and tb1. The enhanced power of this study stemmed from its large genotype panel, improved marker density, and precise RNA-seq data collection from a controlled field setting. Compared to earlier TWAS efforts, the expanded dataset yielded stronger, independent transcript–trait associations. The ability to reuse transcriptomic data across environments and tissues presents an opportunity to accelerate gene discovery in sorghum. These findings lay a genomic foundation for breeding climate-resilient sorghum varieties with optimized flowering times across latitudes.
SorghumBase examples:
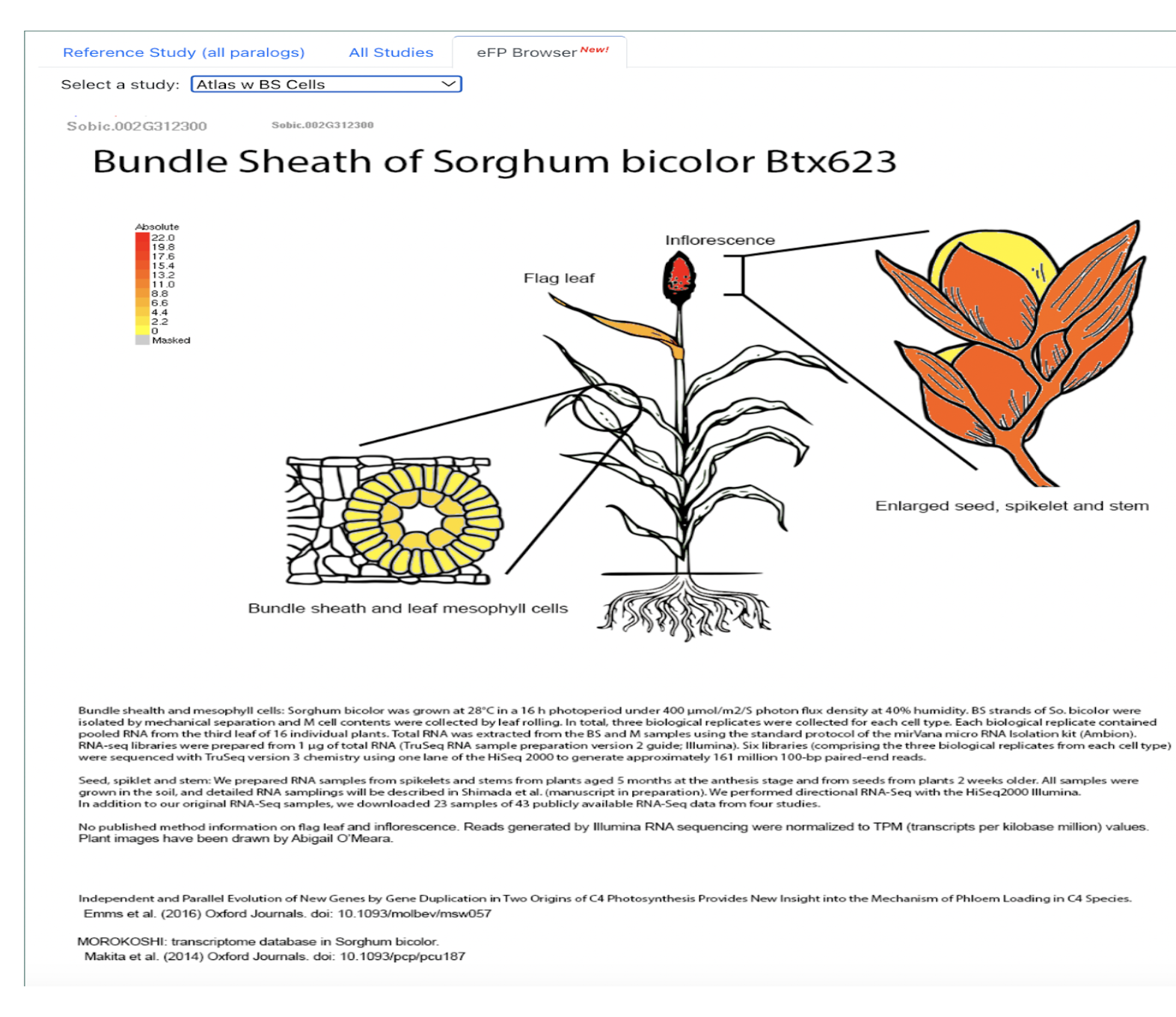
UseCase 1: SbSPB19 (Sobic.002G312300), the gene whose expression was most strongly associated with flowering in this study. Search with gene name Sobic.002G312300 in sorghumbase.org returned hits in both BTx623 assembly v5 and v3. Since most of the annotations are on v3, we selected the v3 gene result.
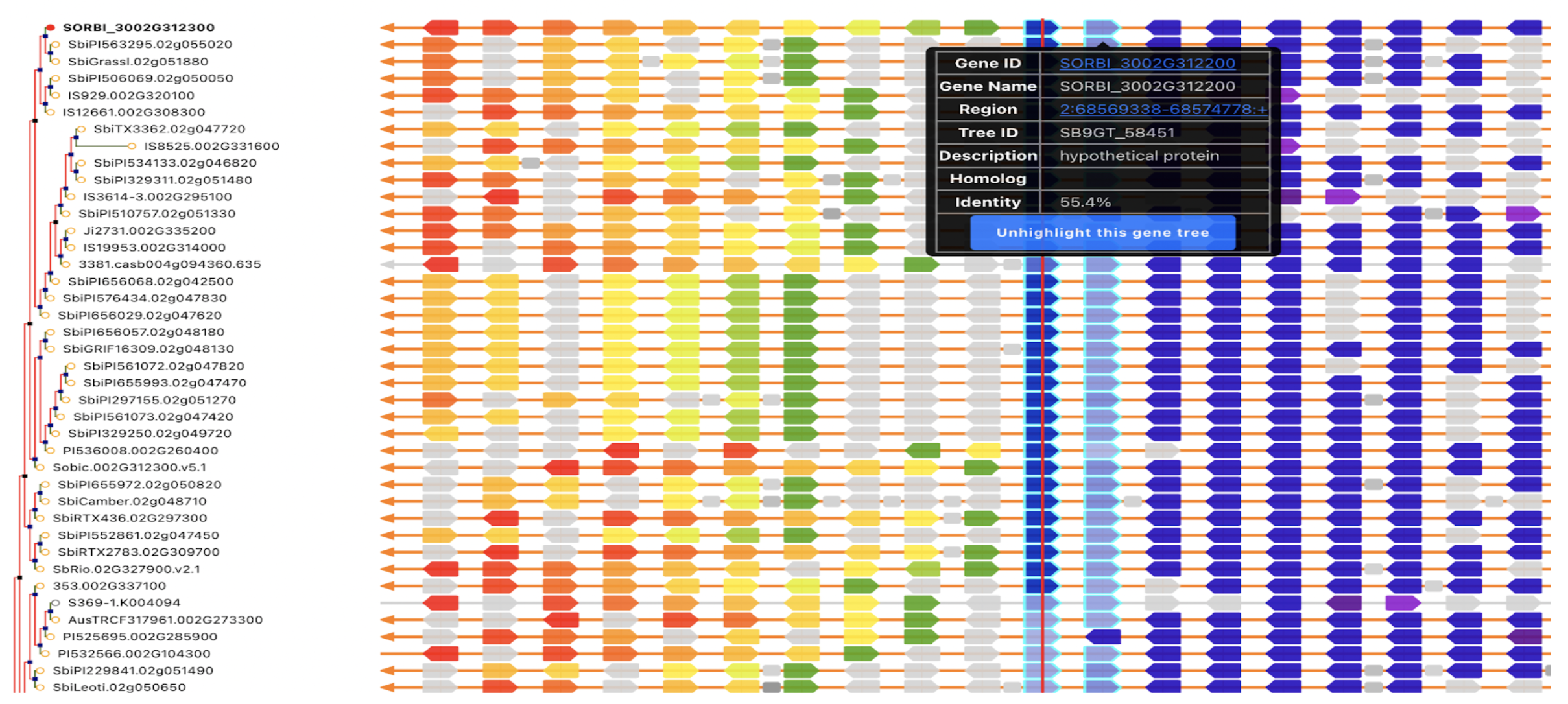
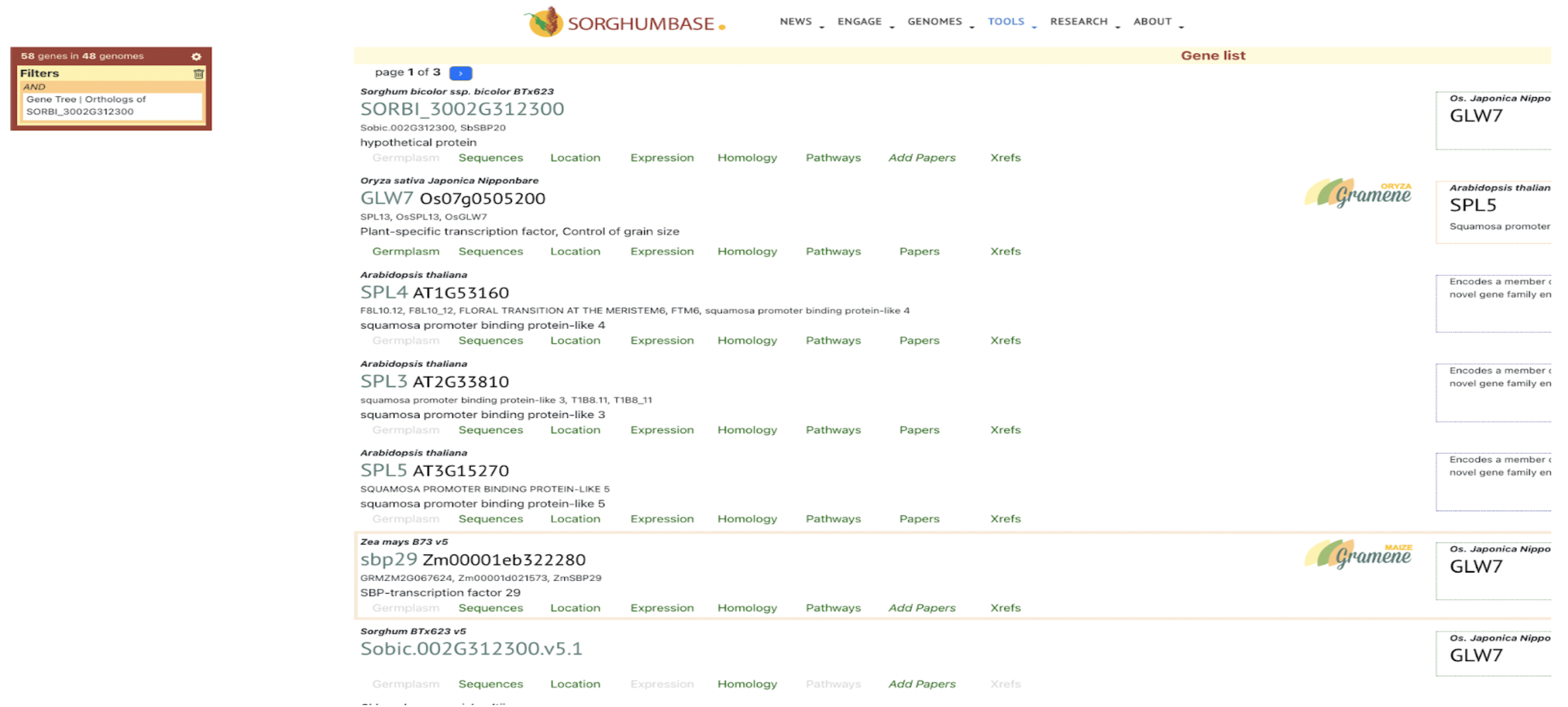
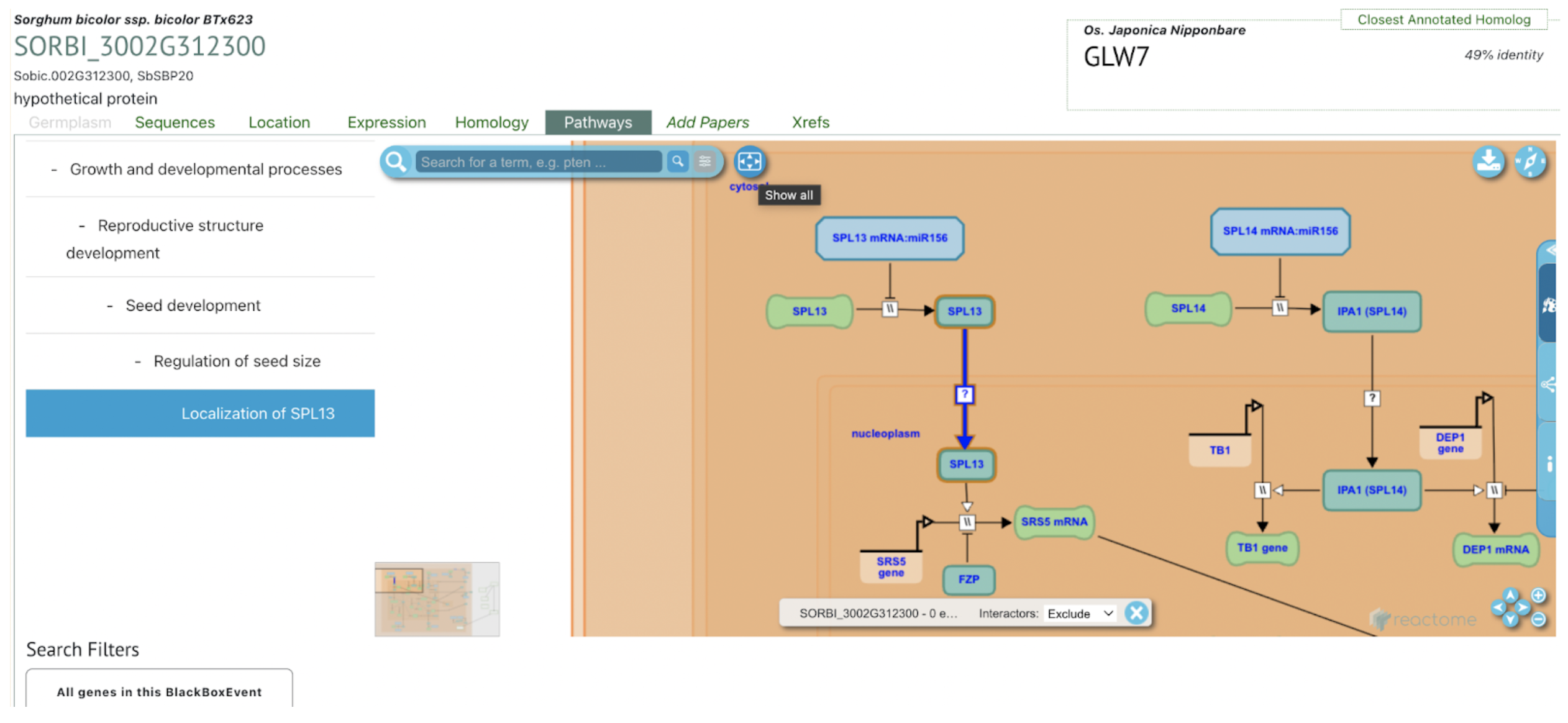
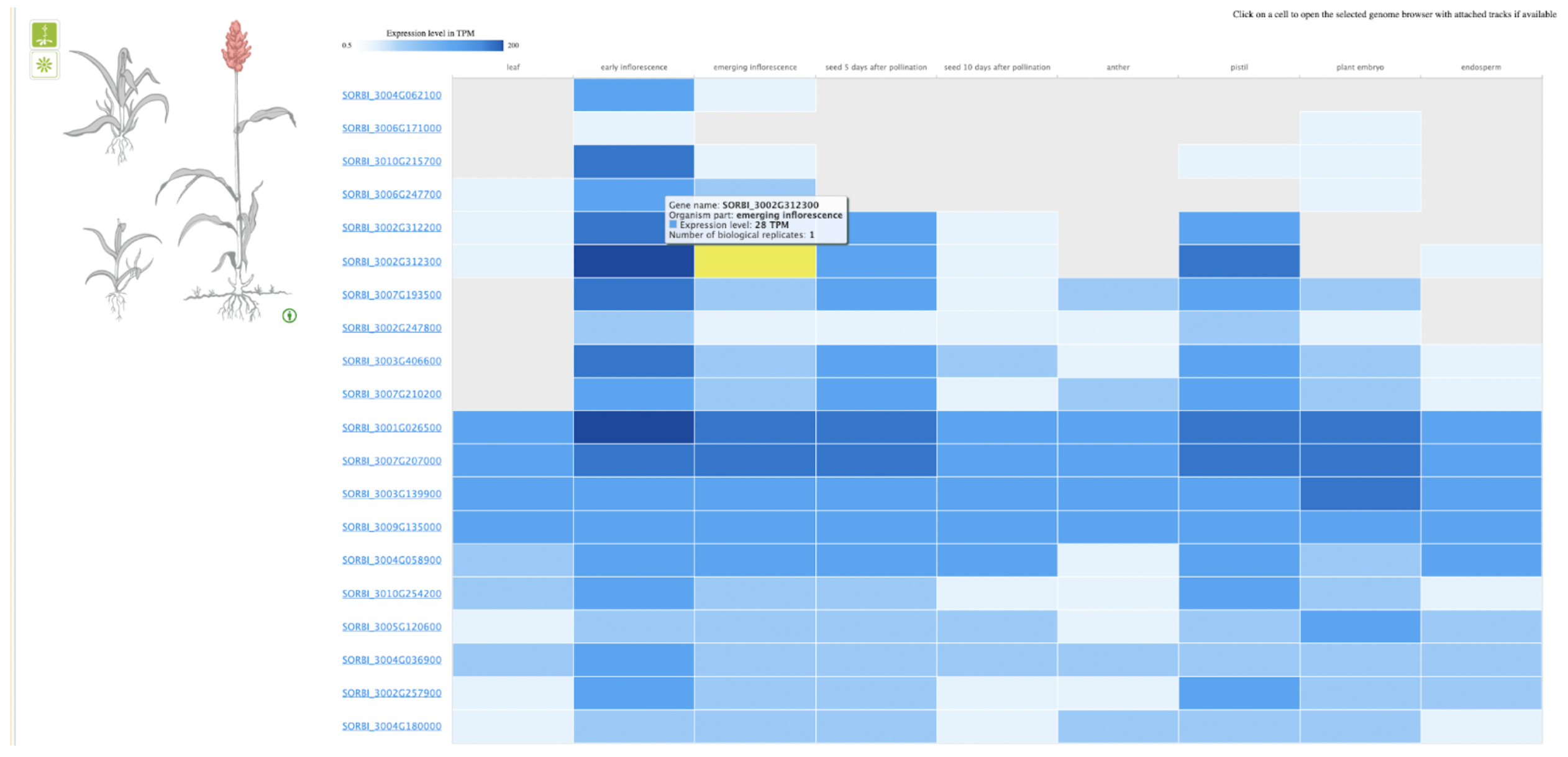
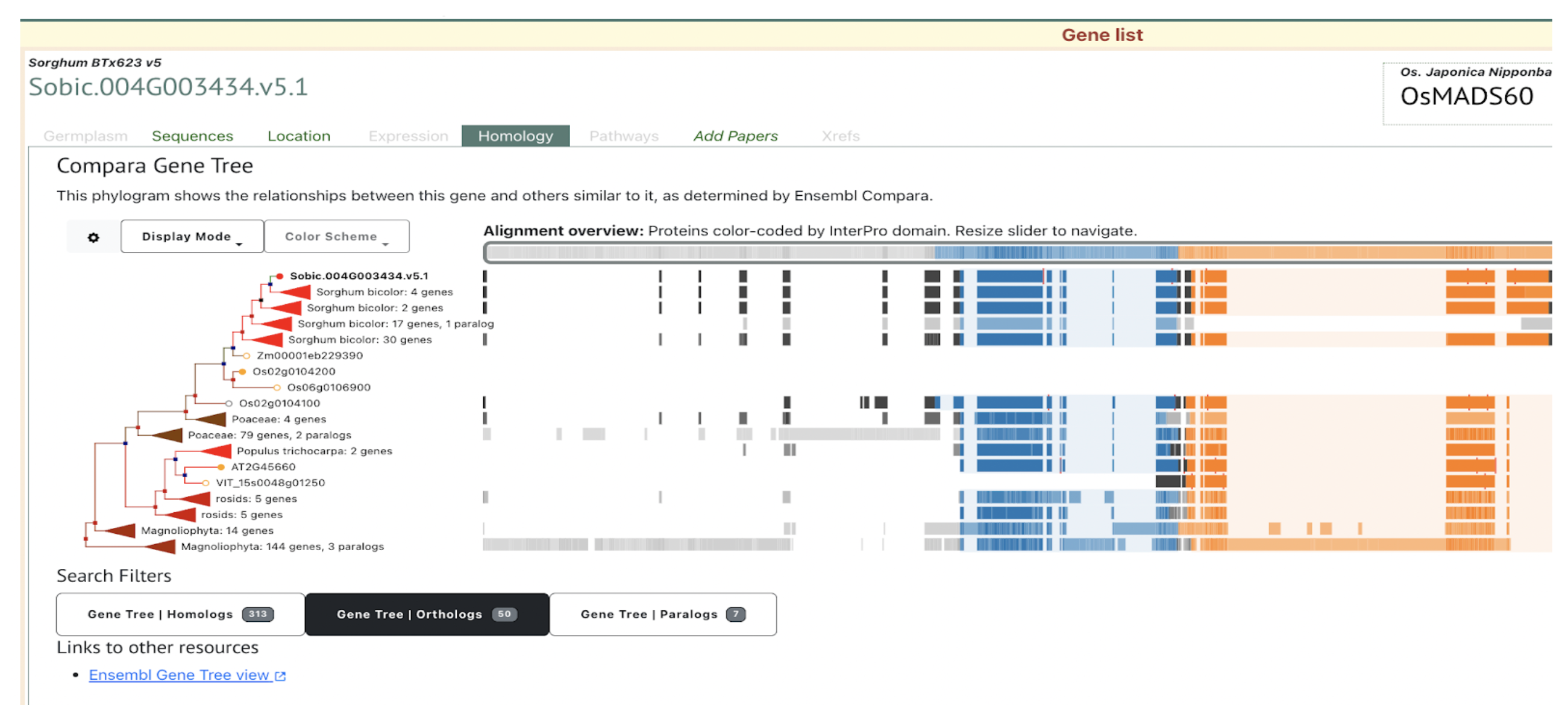
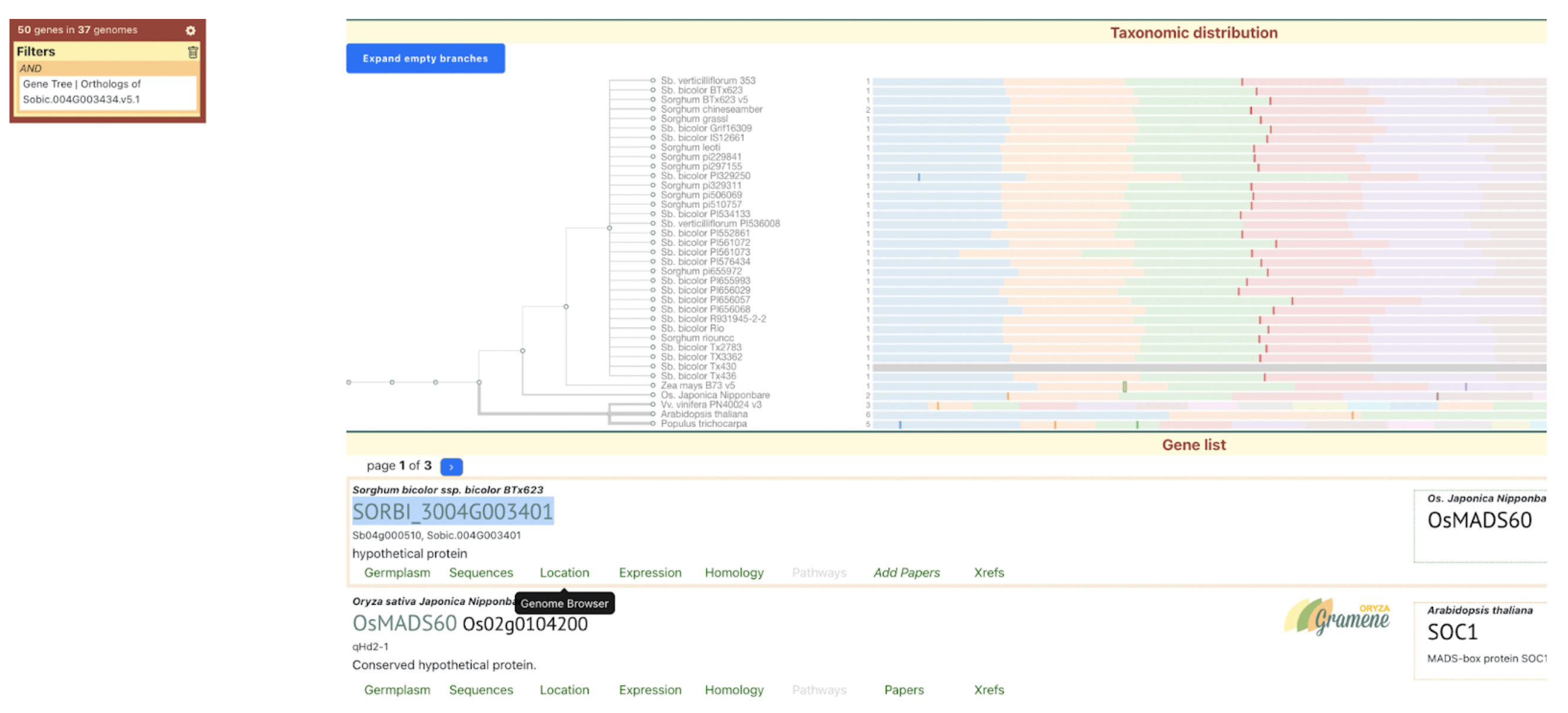
UseCase 2: Gene Sobic.004G003434 (SbMADS31) one of the MADS box genes significantly associated with variation in sorghum flowering time. It represents one of the three lineages from the rice and sorghum common ancestor, and is located in a conserved syntenic region with equivalent intron/exon architecture in rice gene LOC_Os02g01355 (Os02g0104100 is the IRGSPv1 gene name as shown in Figure 2D); however, this conserved gene is apparently completely absent from the maize genome based on both BLAST comparison to annotated maize genes according to the paper, this is confirmed with sorghum multiple species alignment as shown in Fig 2C.


Reference:
Mangal H, Linders K, Turkus J, Shrestha N, Long B, Cebert E, Kuang X, Torres-Rodriguez JV, Schnable JC. Genes and pathways determining flowering time variation in temperate-adapted sorghum. Plant J. 2025 Jun;122(5):e70250. PMID: 40456184. doi: 10.1111/tpj.70250. Read more
Related Project Website:
- Schnable Lab at the University of Nebraska-Lincoln: https://schnablelab.org/
