Ding et al. identified SbC1, an R2R3-MYB transcription factor, as a key regulator of anthocyanin biosynthesis in sorghum coleoptiles, highlighting its role in pigmentation, stress tolerance, and potential applications in crop improvement.
Sorghum’s diverse pigmentation—ranging from coleoptile to grain—derives from a variety of flavonoids such as anthocyanins and tannins, which contribute not only to plant aesthetics but also to stress resistance, grain quality, and bird deterrence. Despite the importance of these compounds, relatively few genes regulating their biosynthesis have been fully characterized. Scientists from Guizhou Academy of Agricultural Sciences and collaborating institutions used genome-wide association studies (GWAS), virus-induced gene silencing (VIGS), and transcriptome analysis to identify SbC1, an R2R3-MYB transcription factor, as a key regulator of anthocyanin biosynthesis in sorghum coleoptiles. Functional validation through heterologous expression in rice and interaction assays confirmed that SbC1 activates key structural genes such as F3H, DFR, ANS, and UFGT. SbC1 interacts with the WD40 protein Tan1 but not with the bHLH protein Tan2, which is grain-specific, indicating a distinct regulatory complex for coleoptile pigmentation. Sequence variation in SbC1 alleles affects anthocyanin accumulation and may have been subject to selection during domestication, especially in Chinese sorghum lines.
These findings highlight SbC1‘s potential utility in breeding programs aiming to improve sorghum resilience to abiotic stress. Anthocyanins accumulated in coleoptiles form a flavonoid-rich barrier that enhances seedling survival under drought and oxidative stress. Comparative analyses with orthologs in maize, rice, and wheat suggest a conserved yet tissue-specific role for C1-like MYB genes across cereals, while also raising the possibility of functional divergence. The study further reveals that the canonical MYB-bHLH-WD40 (MBW) complex may be more variable than previously thought in sorghum, calling for further identification of bHLH partners like SbPLSH1. Overall, this research lays a molecular foundation for targeted manipulation of pigmentation traits to enhance crop performance under stress conditions.
SorghumBase examples:
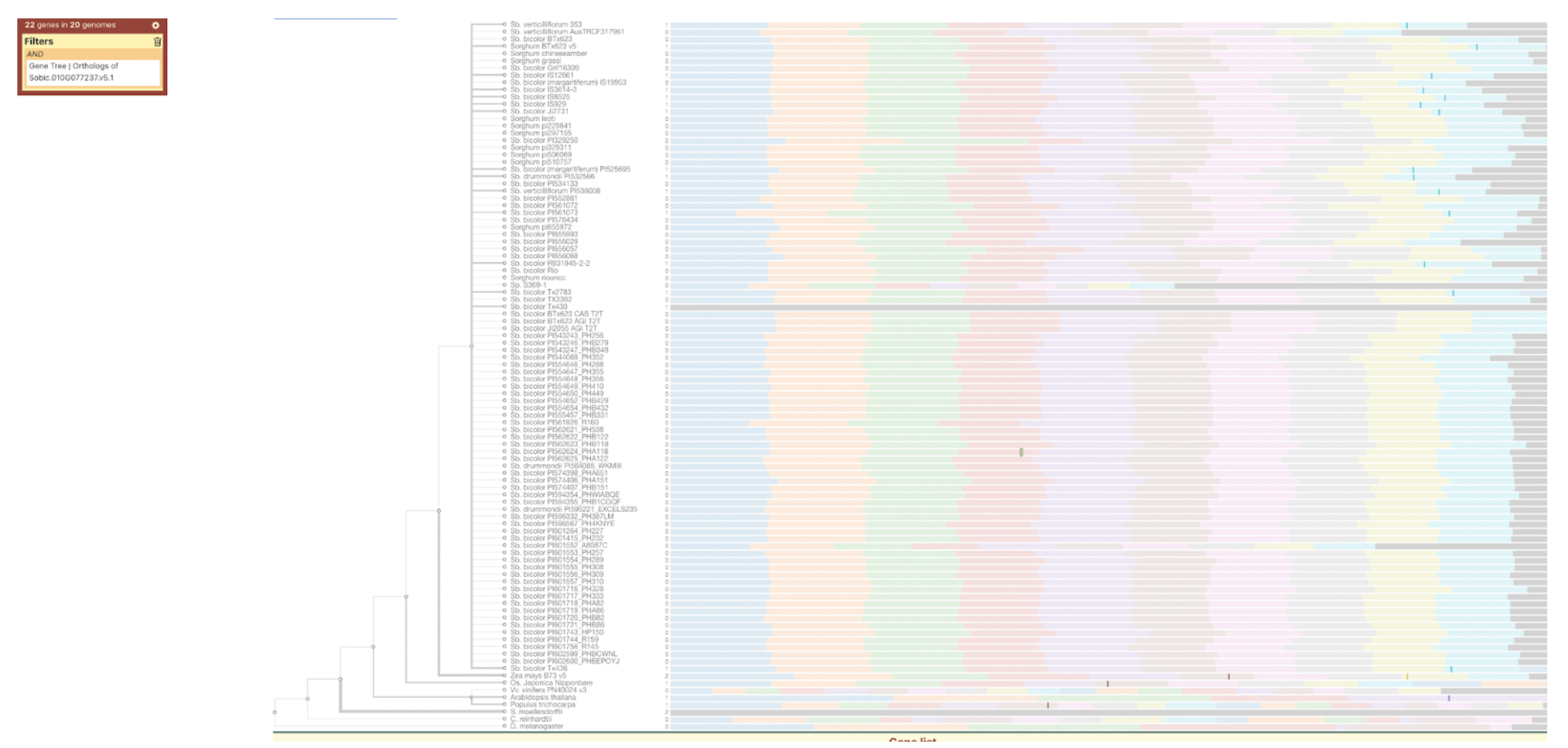
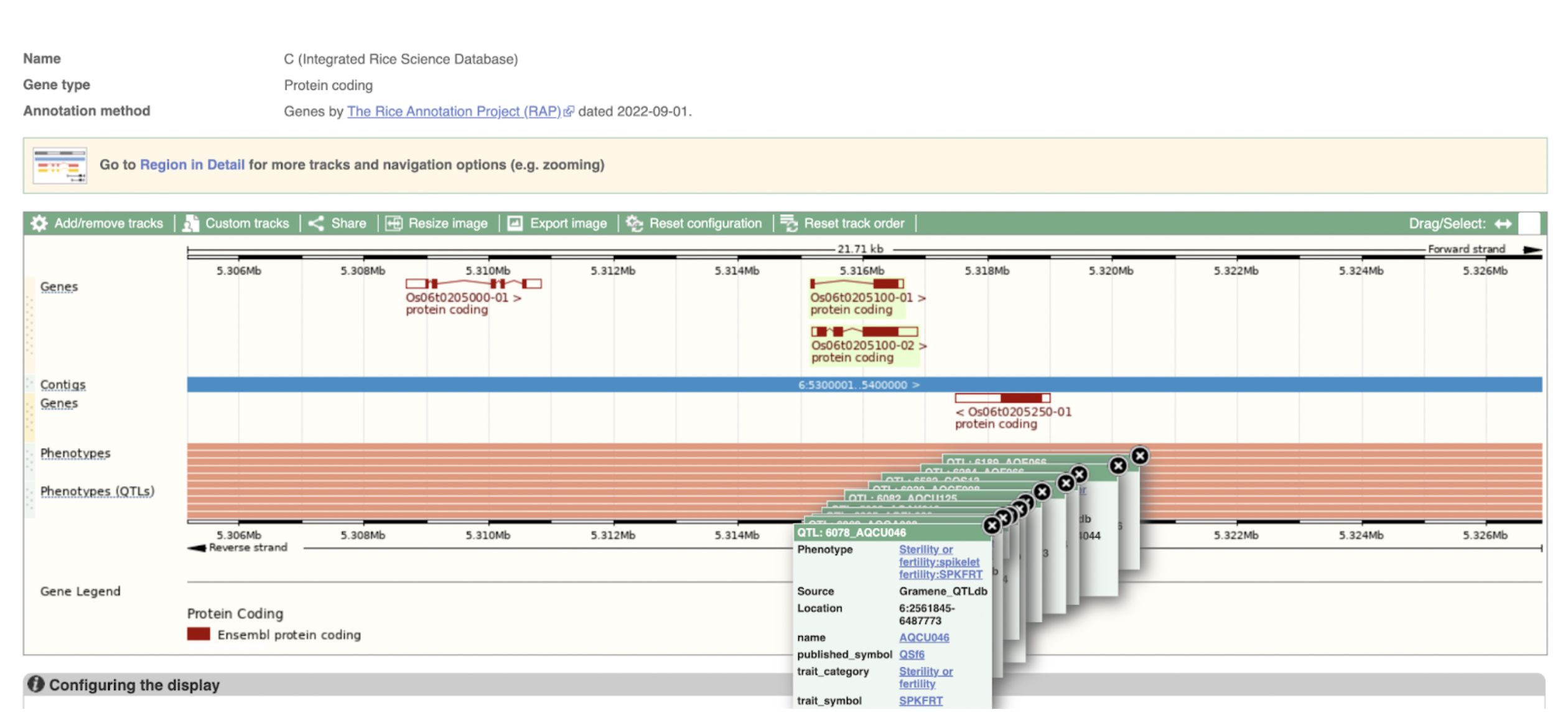
Figure 2A
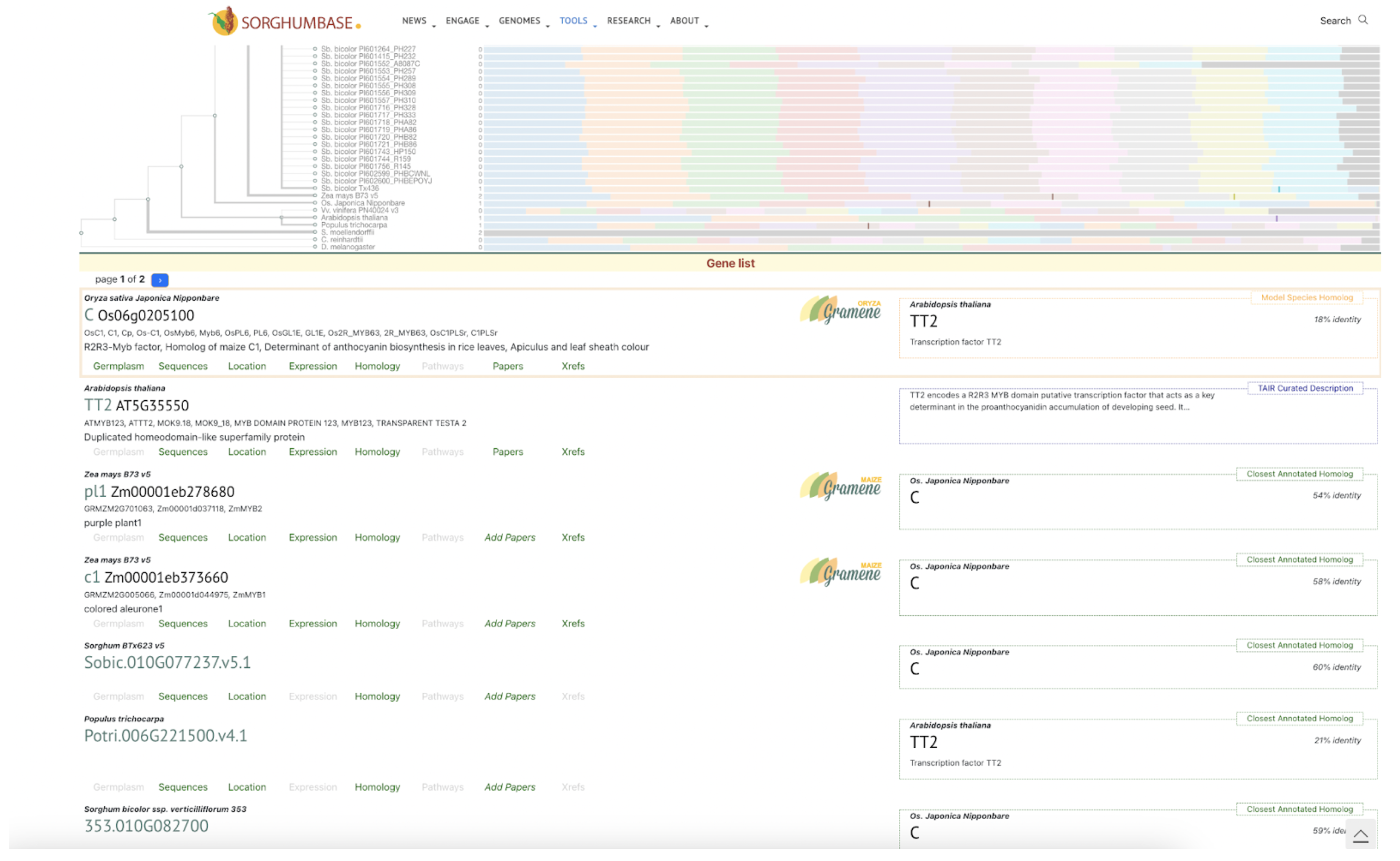
Figure 2B
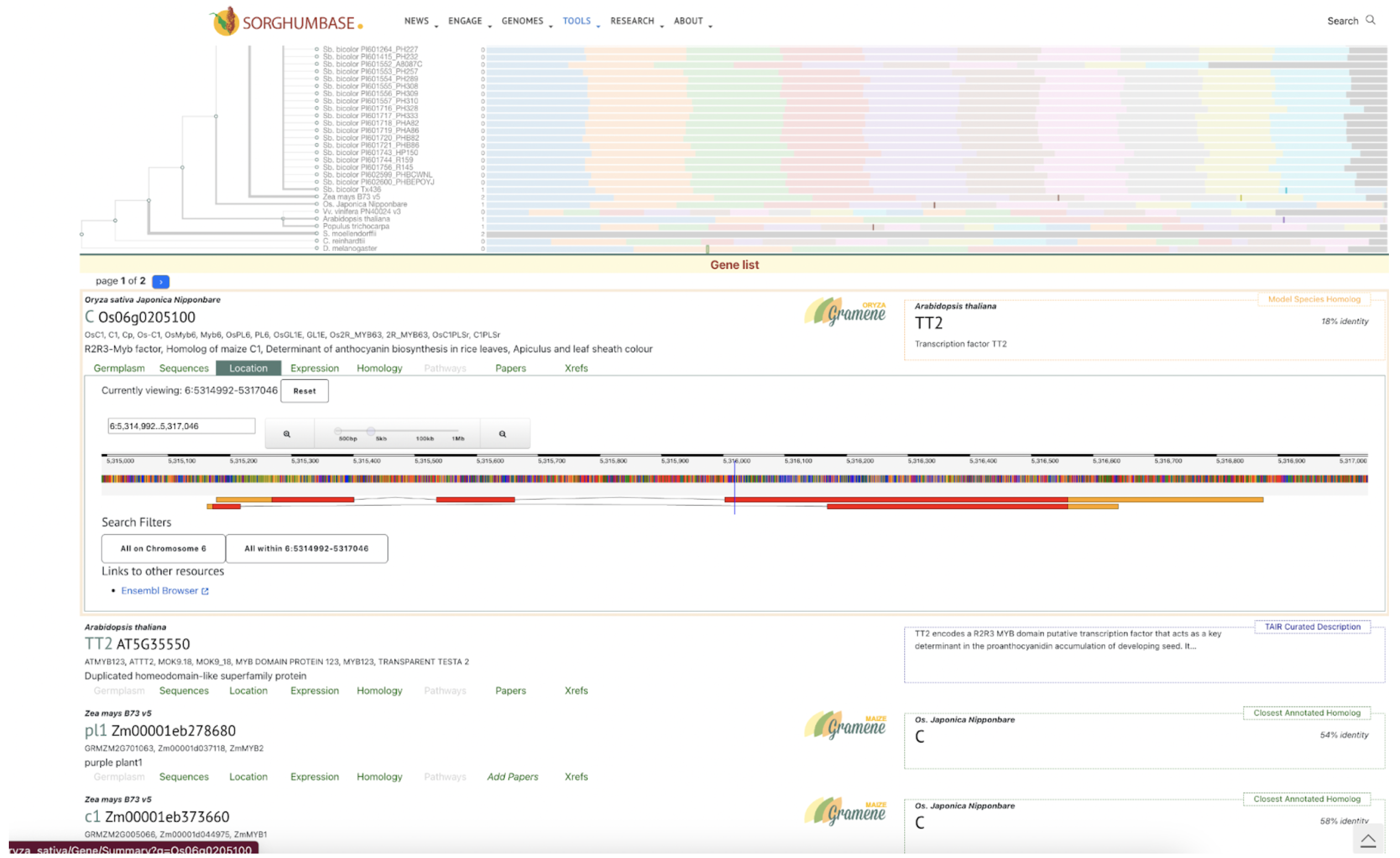
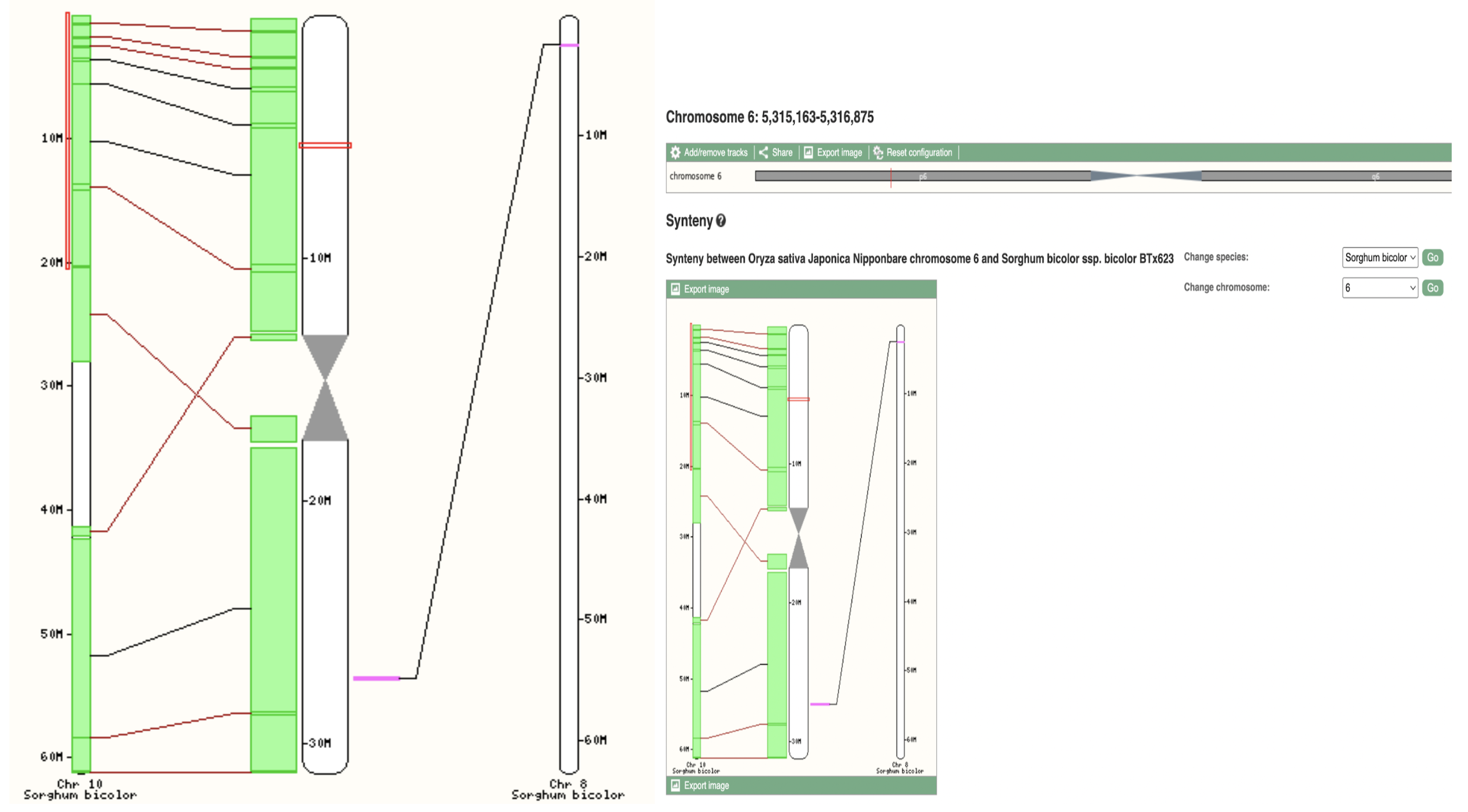
Figure 3A
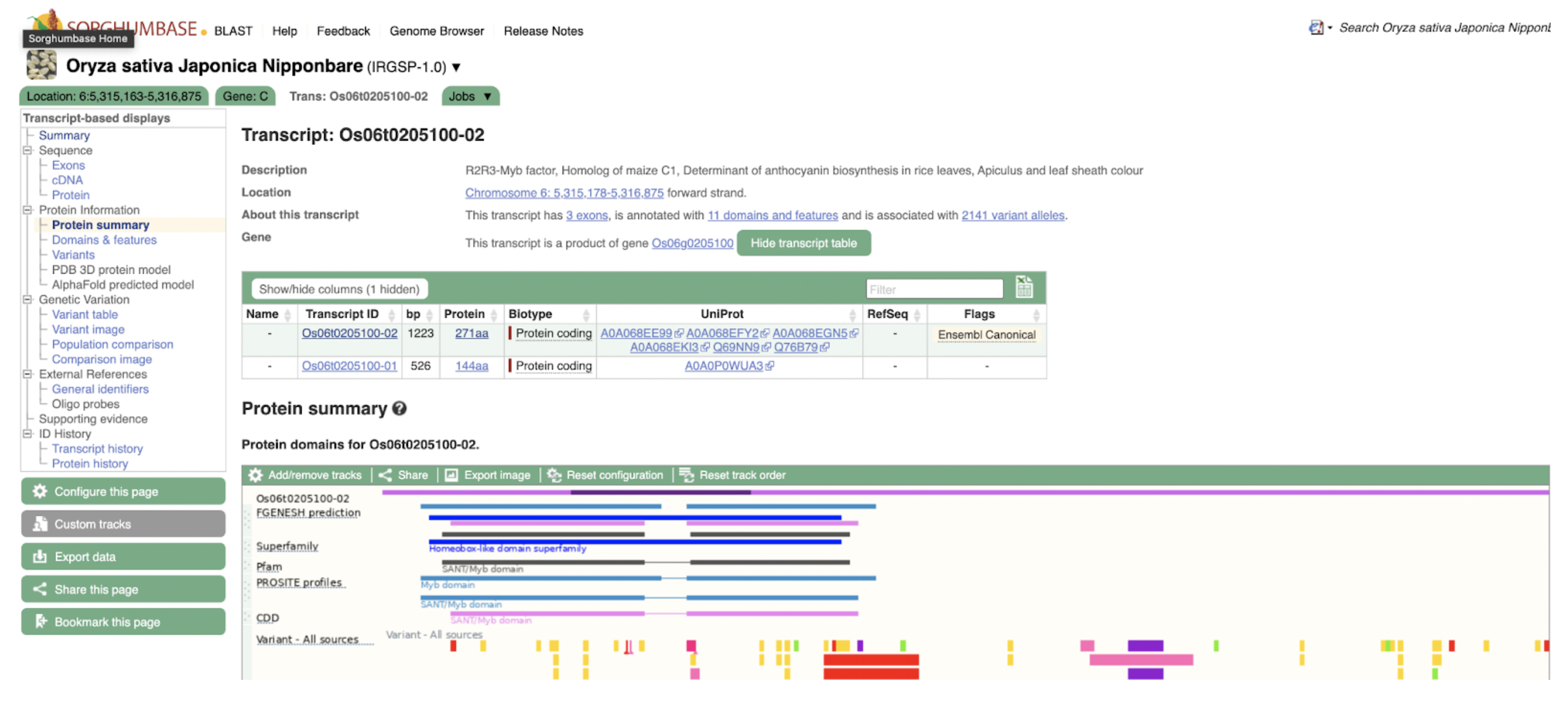
Figure 3B
Reference:
Ding Y, Wang R, He J, Xu J, Cao N, Du J, Li W, Gao X, Cheng B, Luan J, Li S, Zhang L. SbC1, an R2R3-MYB transcription factor, specifically regulates anthocyanin accumulation in sorghum coleoptiles. Theor Appl Genet. 2025 Jun 10;138(7):143. PMID: 40493252. doi: 10.1007/s00122-025-04930-y. Read more