Liu et al. systematically identified and characterized 53 DIR genes in Sorghum bicolor, revealing their evolutionary diversification, tissue-specific expression, and key roles in lignin biosynthesis and abiotic stress tolerance, with SbDIR39 and SbDIR53 highlighted as promising targets for crop improvement.
Keywords: SbDIR gene family, abiotic stresses, systematic evolution, tandem duplication
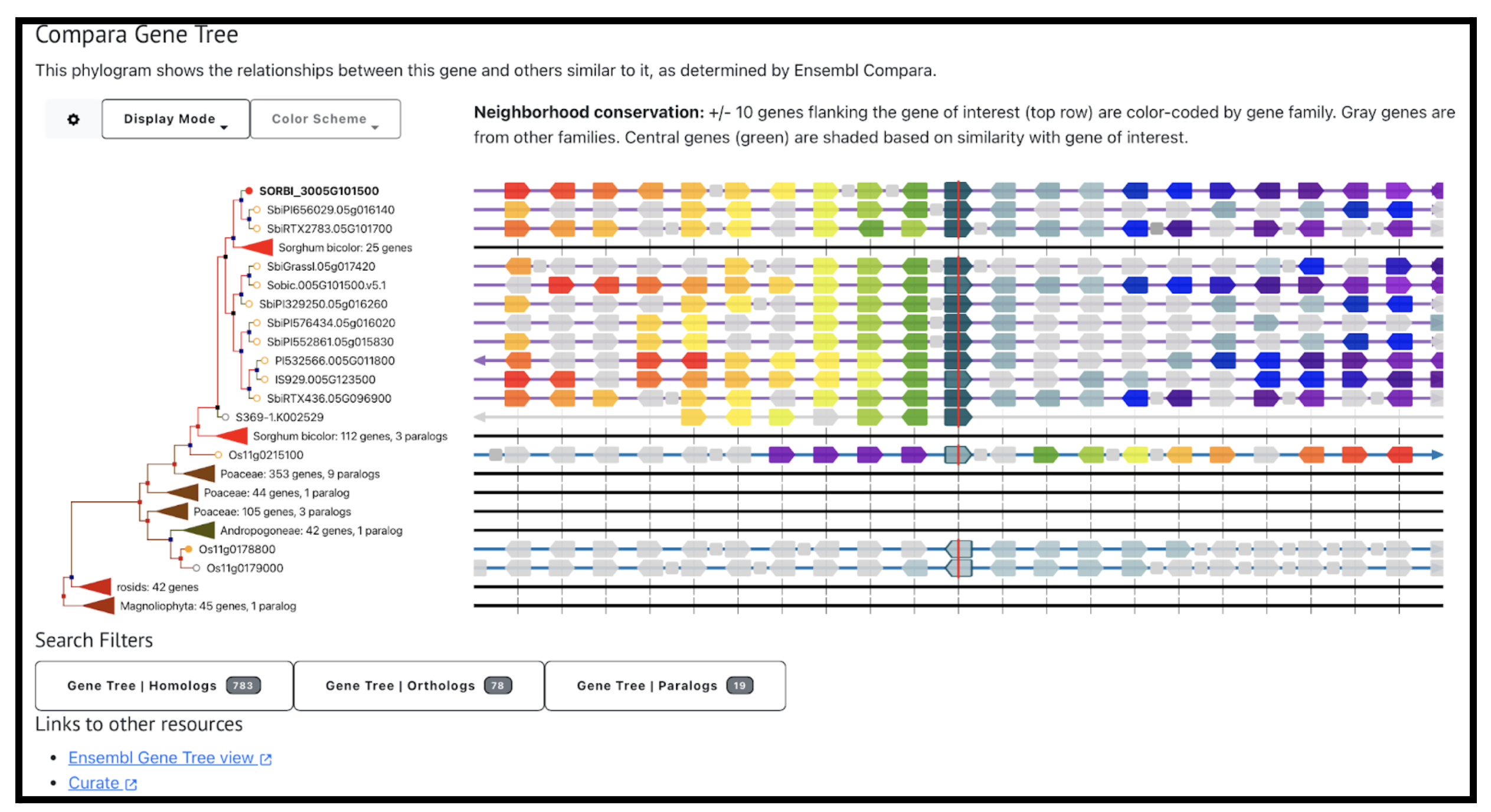
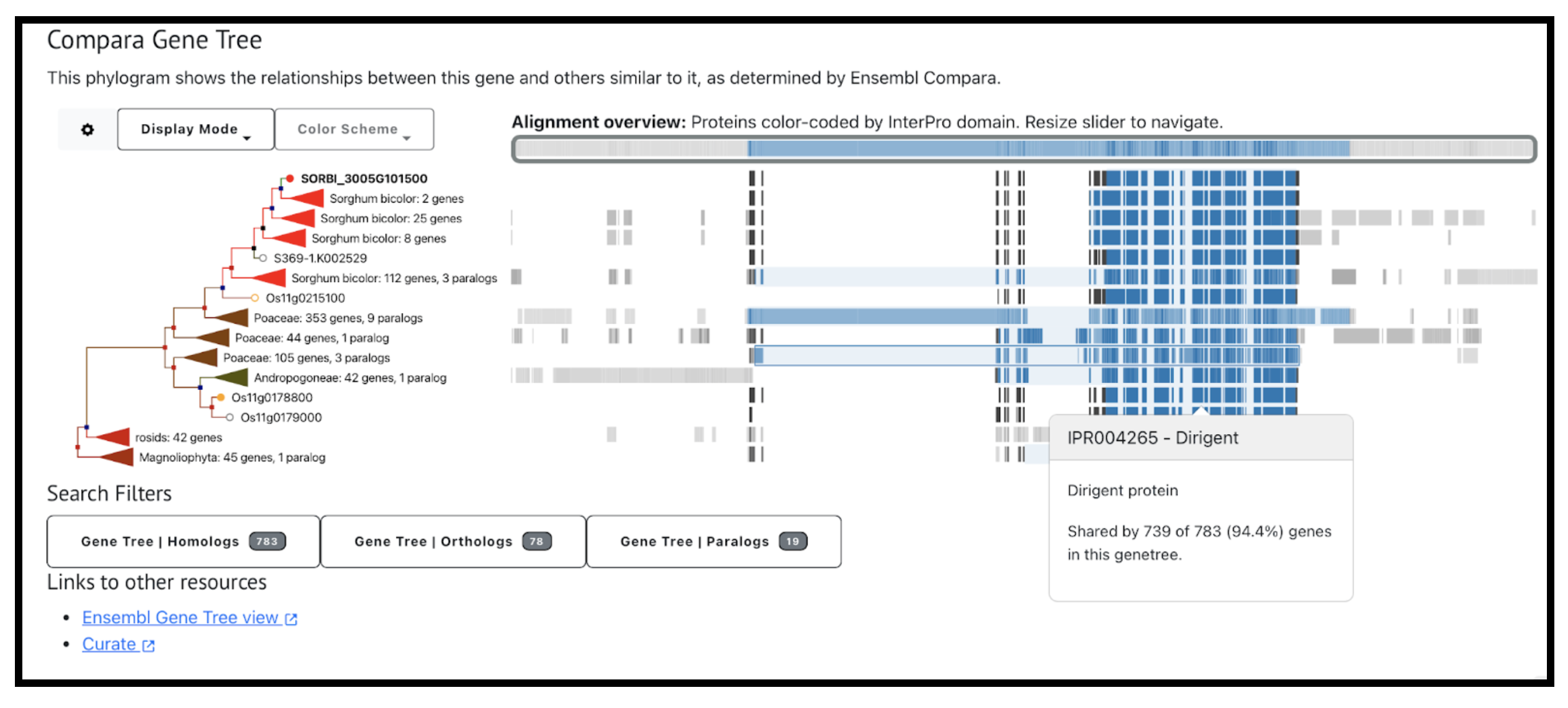
Dirigent (DIR) proteins are a regulatory protein family that play essential roles in secondary metabolism, particularly lignin biosynthesis, stress responses, and developmental processes. While DIR genes have been extensively studied in model plants such as Arabidopsis thaliana and Oryza sativa, their systematic characterization in Sorghum bicolor has been limited despite sorghum’s strong tolerance to drought, salinity, and poor soils. Scientists from Yan’an University identified 53 SbDIR genes classified them into five subfamilies through phylogenetic analysis, with DIR-c and DIR-f uniquely present in monocots. Gene structural variation revealed that intronless DIR-a members may maintain stable expression by avoiding epigenetic silencing, whereas complex exon–intron configurations in DIR-c may facilitate adaptive responses, such as the observed fivefold up-regulation of SbDIR53 under salt stress. Evolutionary analyses further indicated that tandem duplication events were the primary drivers of SbDIR family expansion, consistent with patterns observed in rice, highlighting the importance of duplication in functional diversification.
Functional insights were gained through promoter and expression analyses, which revealed abundant hormone- and stress-responsive cis-elements, particularly ABA-responsive ABRE motifs, in the majority of SbDIR promoters. Transcriptome profiling demonstrated tissue-specific expression, with ten SbDIR genes highly expressed in roots, implicating their role in lignin biosynthesis and deposition to enhance root structural integrity and stress resilience. Several genes, including SbDIR39 and SbDIR53, were strongly induced by salt stress, suggesting their involvement in lignin-mediated reinforcement of cell walls to maintain water and nutrient homeostasis under adverse conditions. These findings support a model in which SbDIR genes regulate lignin deposition and hormonal signaling to optimize sorghum’s adaptation to abiotic stress. Overall, this study establishes a genomic and evolutionary framework for the DIR gene family in sorghum and identifies promising targets, such as SbDIR39 and SbDIR53, for molecular breeding and biotechnological strategies to enhance stress tolerance in cereal crops.
SB Examples:
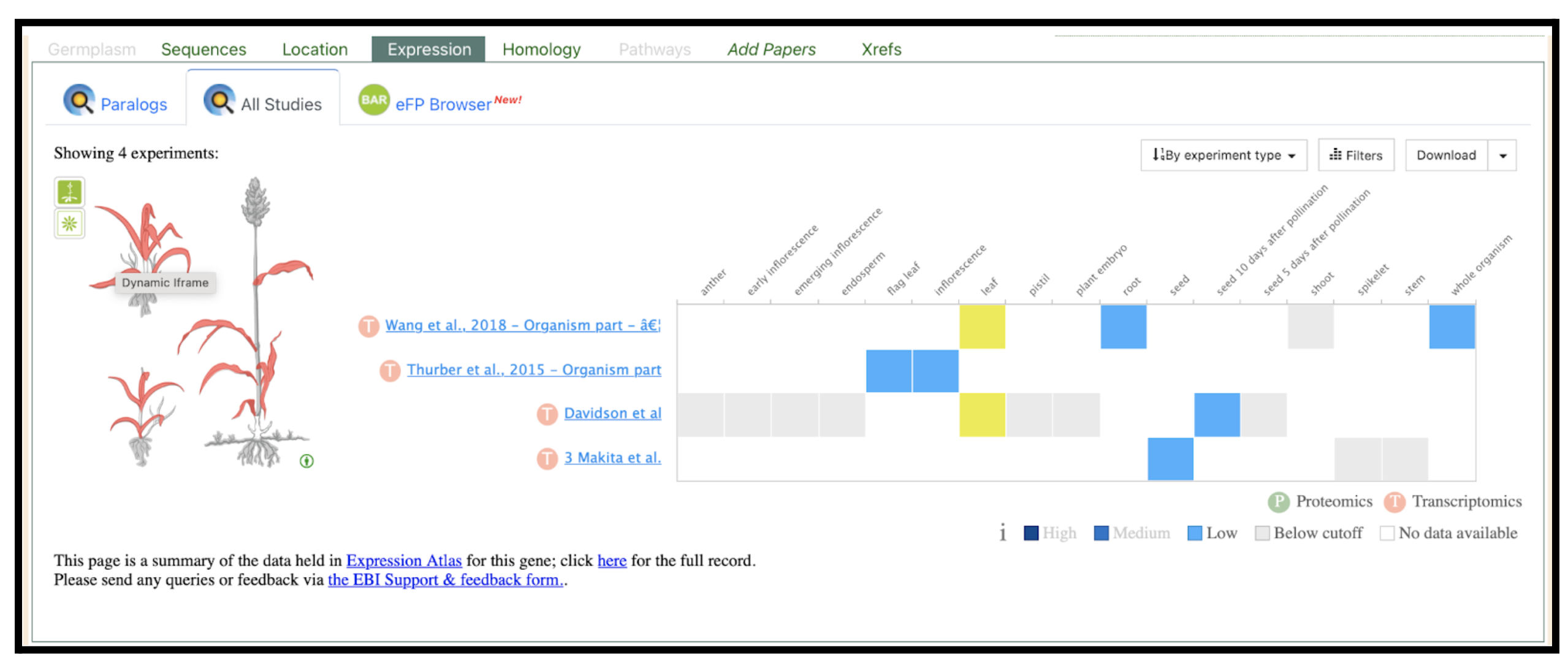
Expression data compiled from various transcriptomics studies reveal tissue-specific expression patterns. Highlighted is the expression patterns in leaves in two studies and indicated by the authors Sobic.005G101500.2, also displayed an expression in leaf when sampling 4 leaves samples. Based on the figure the color intensity reflects relative expression levels (dark = high, medium blue = moderate, light blue = low), while gray indicates below cutoff and white indicates no available data.
Liu S, Jing T, Liang S, Wang H, Guo X, Ma Q, Wang J, Wang K, He X, Zhao H, Jiang W, Zhang X. Genome-Wide Identification of the Dirigent Gene Family and Expression Pattern Analysis Under Drought and Salt Stresses of Sorghum bicolor (L.). Genes (Basel). 2025 Aug 19;16(8):973. PMID: 40870021. doi: 10.3390/genes16080973. Read more