Machine learning–driven GWAS reveals novel candidate genes and pathways underlying the complex, polygenic architecture of grain mold resistance in sorghum.
Plant diseases account for an estimated 10% loss of global food production each year, intensifying food insecurity for over 800 million people. In sorghum, grain mold represents a particularly severe threat, caused by a diverse and variable complex of fungal pathogens that reduce both yield and grain quality while producing harmful mycotoxins. Traditional breeding for resistance has been hindered by the polygenic nature of grain mold resistance, the genetic uniformity of many modern cultivars, and the significant influence of environmental conditions on disease expression. While previous genome-wide association studies (GWAS) have identified loci linked to resistance, their reliance on single-marker linear models limits the ability to capture gene–gene and gene–environment interactions that underlie this complex trait. To address these limitations, ensemble machine learning (ML) methods, such as Boosted Trees and Bootstrap Forests, provide a more powerful framework for modeling nonlinear relationships across high-dimensional genetic data.
In this study, ML-driven GWAS was applied to a sorghum association panel, integrating diverse phenotypic representations of grain mold response. This approach uncovered a suite of candidate genes and genomic regions, including Sobic.005G141700, Sobic.003G329100, and Sobic.002G270800, many of which were not detected by traditional GWAS. Functional predictions suggest roles in pathogen recognition, cellular stress responses, mitochondrial function, and DNA repair, highlighting the multifaceted defense mechanisms contributing to resistance. Gene ontology enrichment further supported the involvement of pathways related to genome stability, redox homeostasis, and immune signaling. By leveraging ensemble ML methods, this work not only refines our understanding of the genetic architecture of grain mold resistance but also provides valuable molecular targets for marker-assisted and genomic selection. These findings demonstrate the potential of ensemble ML-driven approaches for dissecting polygenic traits and advancing the development of durable disease resistance in crops.
SorghumBase Examples:
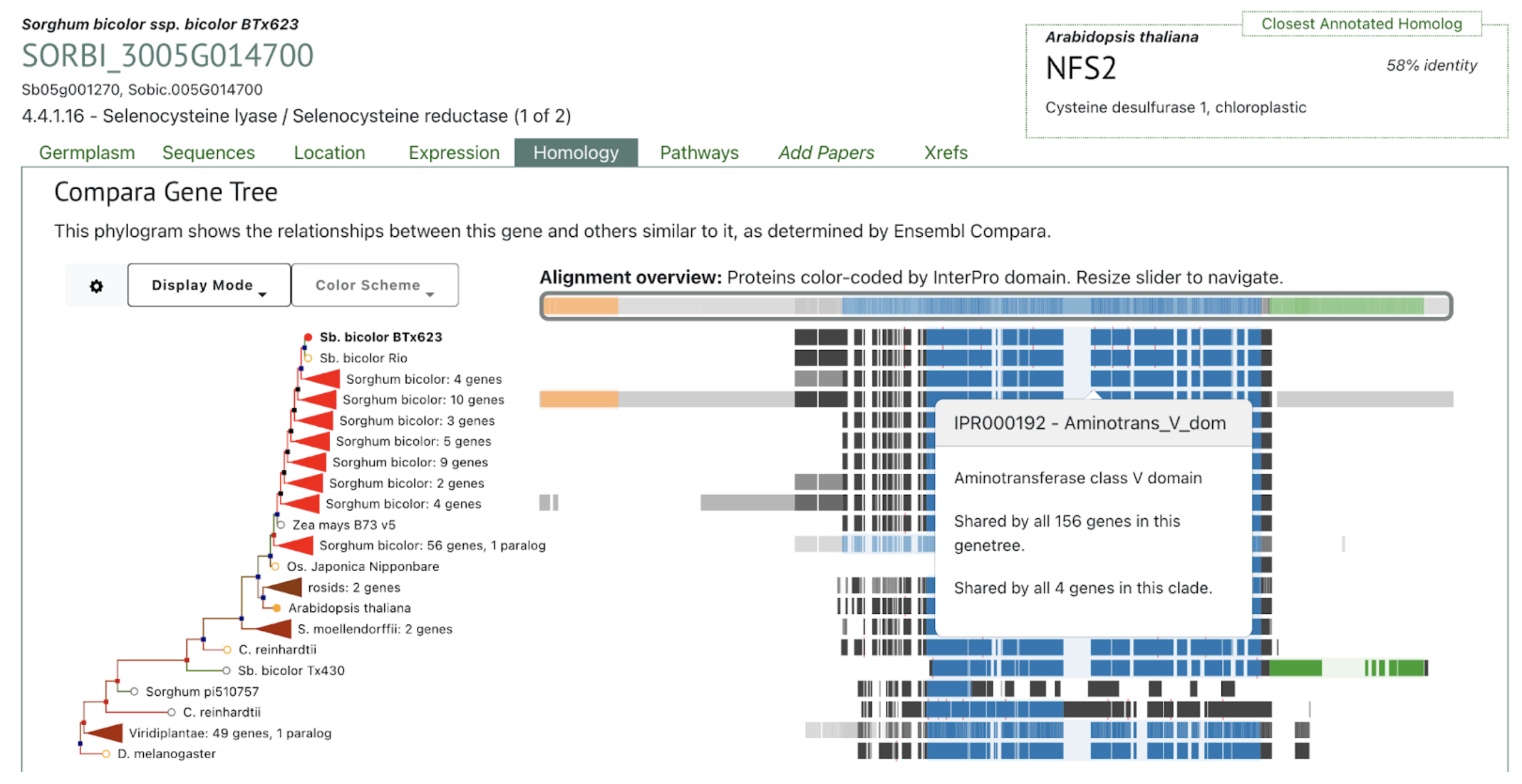
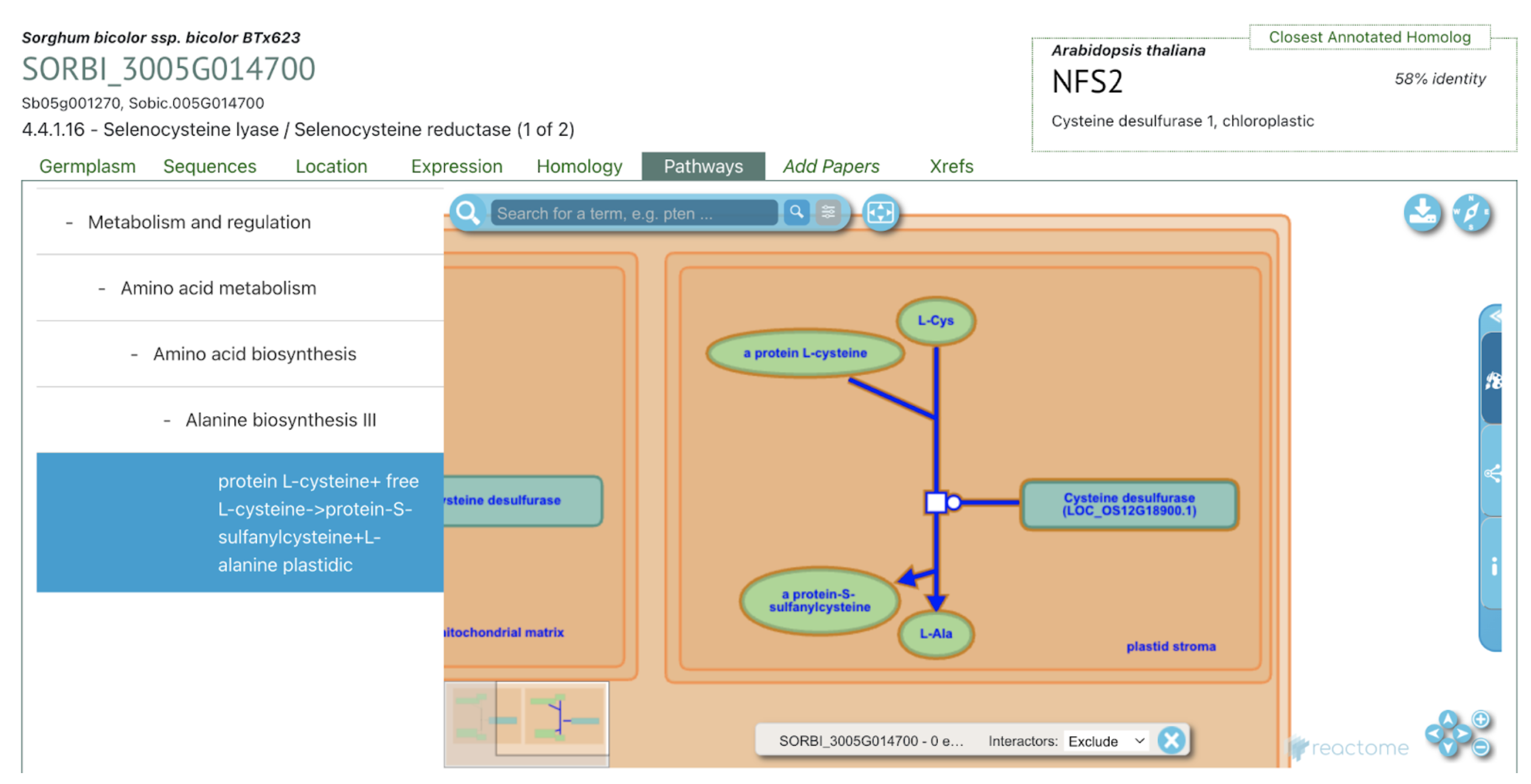
Reference:
Ahn E, Prom LK, Park S, Lee D, Bhatt J, Ellur V, Lim S, Jang JH, Lakshman D, Magill C. Machine learning reveals complex genetics of fungal resistance in sorghum grain mold. Heredity (Edinb). 2025 Aug;134(8):485-499. PMID: 40684039. doi: 10.1038/s41437-025-00783-9. Read more
Related Project Websites:
- Clint Magill’s lab at Texas A&M University: https://sites.google.com/tamu.edu/magill-lab-tamu/home