Mutations in the sorghum Waxy (GBSS) gene reduce amylose content and create value-added grain traits, and new genomic resources now enable more efficient breeding of waxy sorghum for food, feed, and biofuel applications.
Keywords: Breeding, Grain quality, Molecular markers, Sorghum, Waxy
This work was performed thanks to our lab’s first federal grant after starting up at the University of Nevada, Reno. We are deeply grateful to have received that funding and to Dr. Sarah Sexton-Bowser, who connected me with Richardson Seeds to lead this public-private effort. – Yerka
Mutations in the GRANULE-BOUND STARCH SYNTHASE (GBSS) gene (Sobic.010G022600), commonly referred to as Waxy, reduce amylose content in sorghum endosperm, resulting in a sticky “waxy” texture. While waxy sorghum has long been valued for improved digestibility, rapid ethanol fermentation, and functional food applications, early breeding efforts reported inconsistent grain yields and poor germination, limiting its adoption. The release of near-isogenic waxy/wild-type parent lines in 2015 having no reduction in hybrid yield relative to wild-type hybrids marked a turning point, establishing waxy grain as one of the first value-added sorghum traits in the United States. However, breeding for the Waxy locus has traditionally relied on PCR-based marker-assisted selection (MAS), which is incompatible with next-generation sequencing (NGS) and requires separate genotyping steps during genomic prediction and selection (GP/GS). This dual process is both time-intensive and costly. To overcome these challenges, researchers from the University of Nevada, Reno, Texas Tech University, USDA-ARS, Richardson Seeds Ltd. and Nuseed developed new molecular resources—including high-throughput PACE markers and an in silico B.Tx623 wxa genome assembly— to improve read mapping, streamline MAS, and integrate Waxy allele tracking into GP/GS pipelines.
The waxy trait, once considered Mendelian, is now understood to exhibit quantitative variation due to the influence of starch branching enzymes and complex genetic architecture. Notably, genome-wide association studies (GWAS) for amylose content in sorghum have identified significant loci but failed to detect significant SNPs at the Waxy locus, underscoring the need for improved marker systems. Current breeding strategies focus on enhancing agronomic performance of waxy germplasm—including heterosis, stress tolerance, and grain quality—while integrating food and nutrition traits such as minerals, phenolics, fiber, protein, and resistant starch content. Emerging approaches, including genomic prediction (GP), genome-to-phenome (G2P) modeling, and multiomic phenotyping, provide tools to clarify pleiotropic interactions among whole-plant metabolism and seed development, including grain starch, protein, and oil biosynthesis pathways. Together, these genomic and phenomic resources will be bolstered by a modern molecular toolkit to accelerate breeding of waxy sorghum for food, feed, and biofuel applications, while supporting the development of climate-smart sorghum varieties with improved yield, nutritional value, and adaptability.
SorghumBase Examples:
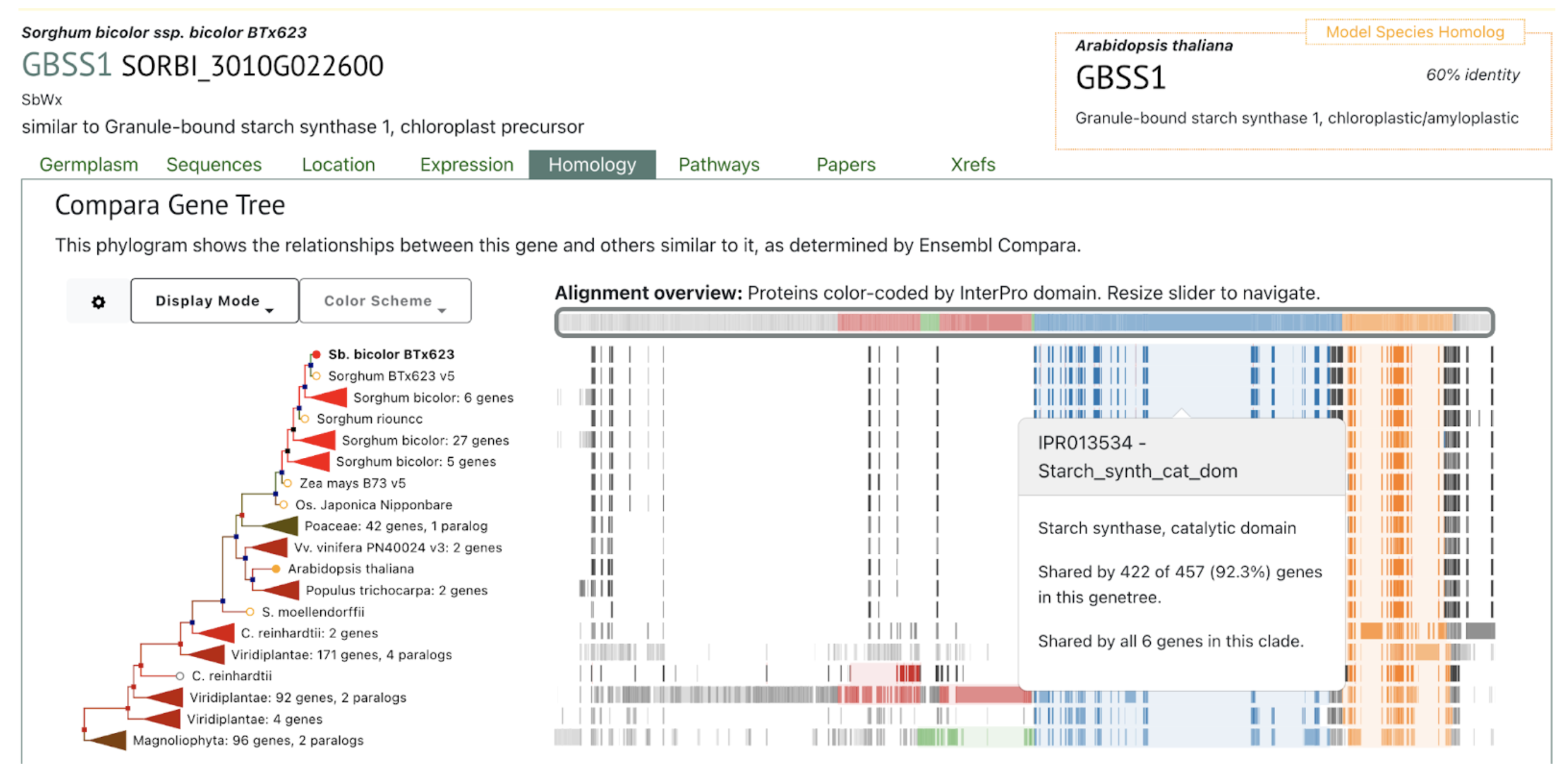
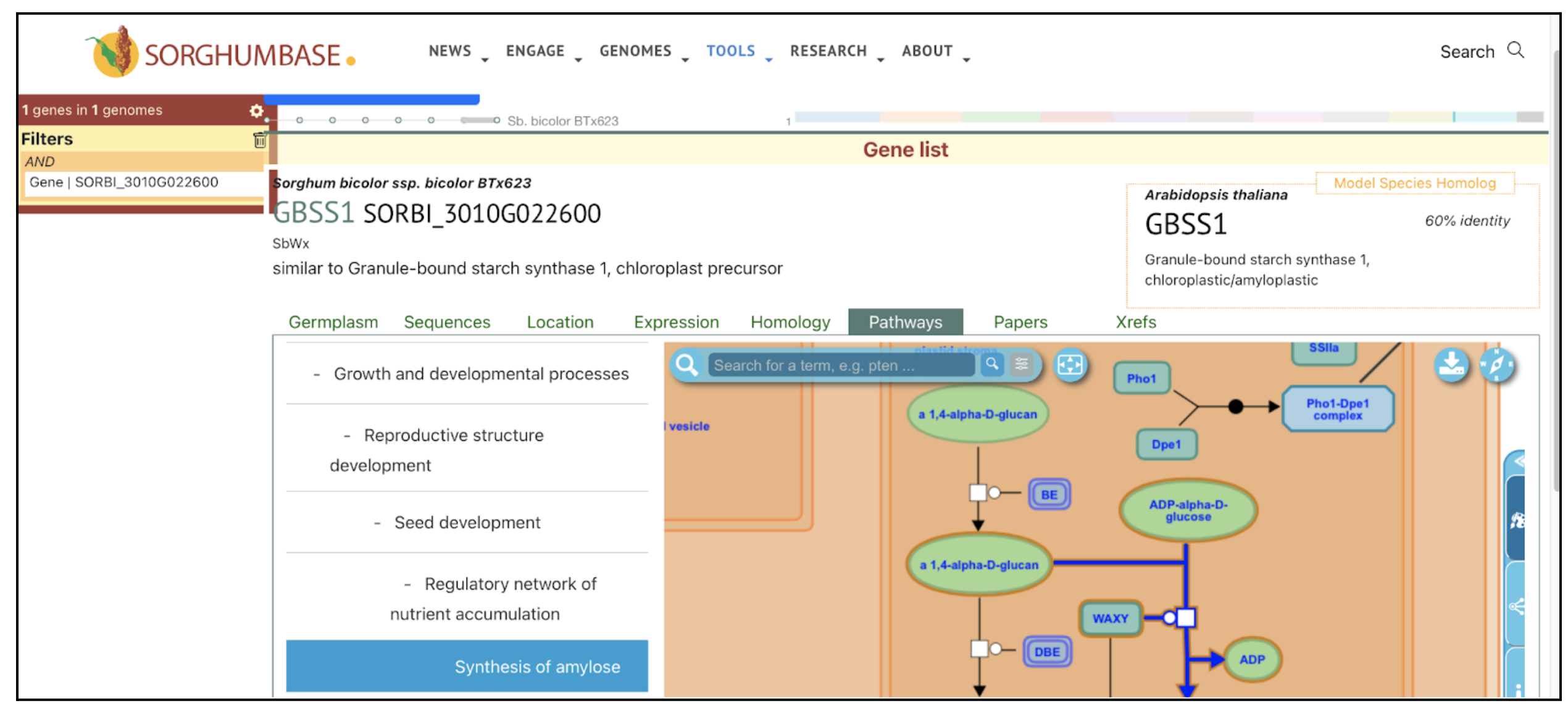
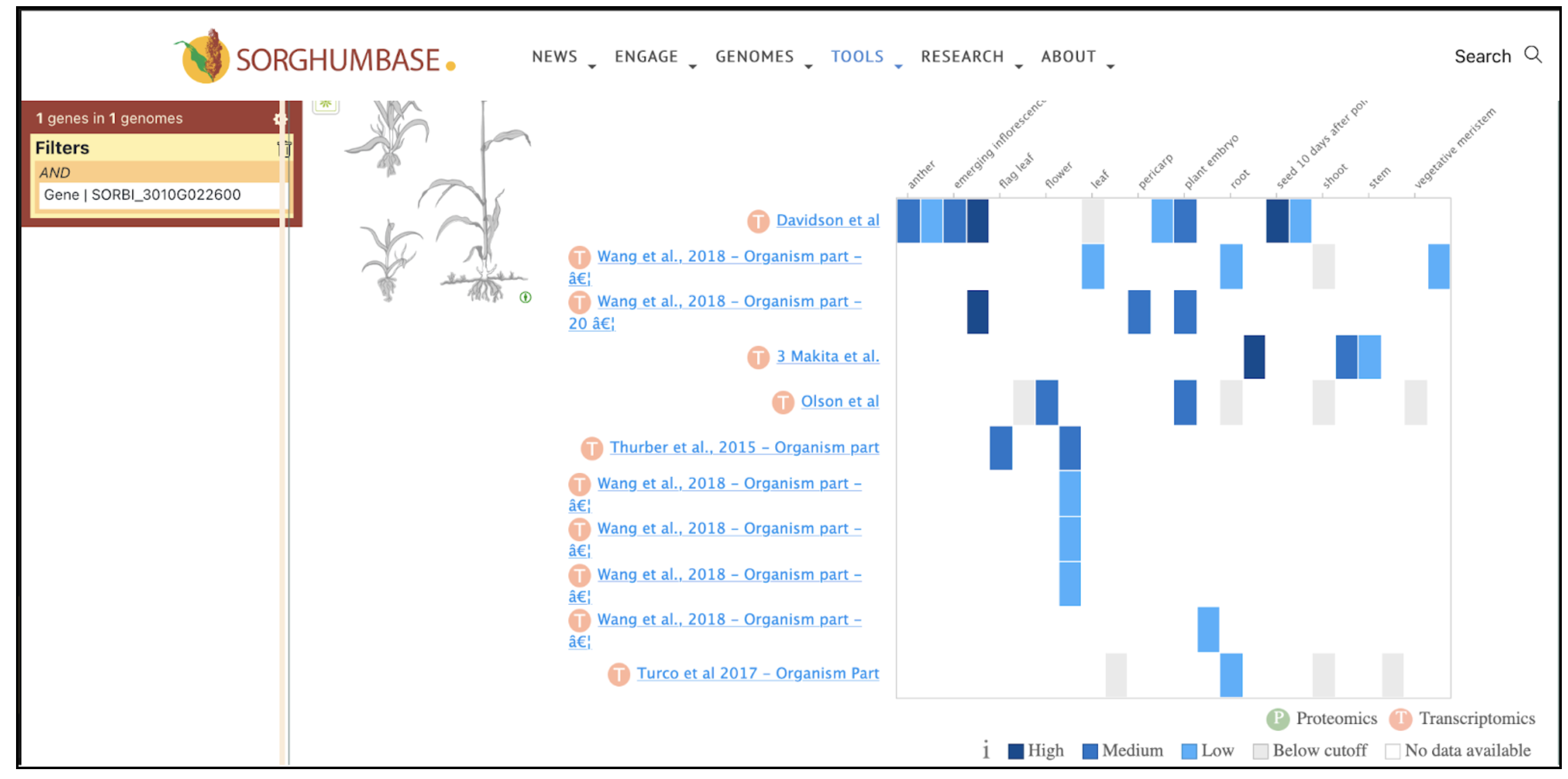
Reference:
Yerka MK, Liu Z, Bean S, Nigam D, Hayes C, Druetto D, Krishnamoorthy G, Meiwes S, Cucit G, Patil GB, Jiao Y. An updated molecular toolkit for genomics-assisted breeding of waxy sorghum [Sorghum bicolor (L.) Moench]. J Appl Genet. 2025 Aug 11. PMID: 40784926. doi: 10.1007/s13353-025-00993-1. Read more
Related Project Websites:
- Yinping Jiao’s Lab at Texas Tech University: https://www.depts.ttu.edu/igcast/Staff/jiao_lab.php
- Institute of Genomics for Crop Abiotic Stress Tolerance (IGCAST) web page: https://www.depts.ttu.edu/igcast/
