A genome-wide association study in sorghum identified 189 QTNs underlying root system architecture, highlighting a complex polygenic basis with candidate genes linked to hormone signaling, flavonoid biosynthesis, and stress adaptation.
Keywords: Multi-locus, Root angle, Root length, Root to shoot ratio and quantitative trait loci
Understanding the genetic basis of root system architecture is fundamental to building climate-resilient agriculture. In sorghum, a crop that sustains millions in drought-prone regions, our findings illuminate the complex and polygenic nature of early root development. By identifying robust QTNs and linking them with key biological pathways including flavonoid biosynthesis, auxin signaling, and stress-response mechanisms. we provide a genomic foundation for harnessing root traits in breeding programs. This research not only validates the power of integrated GWAS approaches but also opens new avenues for discovery through uncharacterized genes. Ultimately, strengthening sorghum root systems means enhancing water-use efficiency, improving yield stability, and securing food systems in the face of climate uncertainty. – Tsehaye
Root system architecture (RSA) is a critical determinant of plant growth, nutrient uptake, and resilience to environmental stress, particularly drought. Sorghum, a staple crop in many arid and semi-arid regions, is increasingly recognized for its drought tolerance, making it a strong model for RSA studies. Scientists from Mekelle University, Tigray Agricultural Research Institute, Jimma University and Ethiopian Institute of Agricultural Research conducted a genome-wide association study (GWAS) to identify genomic regions associated with early-stage root traits such as root length, angle, number, and dry weight. A total of 189 quantitative trait nucleotides (QTNs) were identified across all ten sorghum chromosomes, reflecting the polygenic and trait-specific nature of RSA. The detection of QTNs varied among multi-locus mixed models (MLMs), with approaches incorporating both population structure and kinship (e.g., pKWmEB, pLARmEB) proving particularly effective. Importantly, 38 QTNs were consistently detected across multiple models, providing strong evidence for their reliability. These findings indicate that sorghum RSA is governed by a complex genetic architecture influenced by both genetic variation and environmental interactions.
Further analysis revealed substantial overlap between newly identified QTNs and previously reported quantitative trait loci (QTLs), validating the results and suggesting functional relevance. Several QTNs co-located with genes implicated in flavonoid biosynthesis, auxin signaling, and hormone response pathways—processes known to regulate root growth, branching, and stress adaptation. Notably, chalcone synthase and RNA-binding KH domain proteins emerged as candidate genes with potential roles in root development and environmental response. The prevalence of uncharacterized proteins among co-located genes underscores opportunities for novel gene discovery. Collectively, these results provide a valuable genomic resource for sorghum improvement, enabling marker-assisted selection and targeted breeding strategies aimed at enhancing yield, water-use efficiency, and stress tolerance through optimized RSA.
SorghumBase Examples:
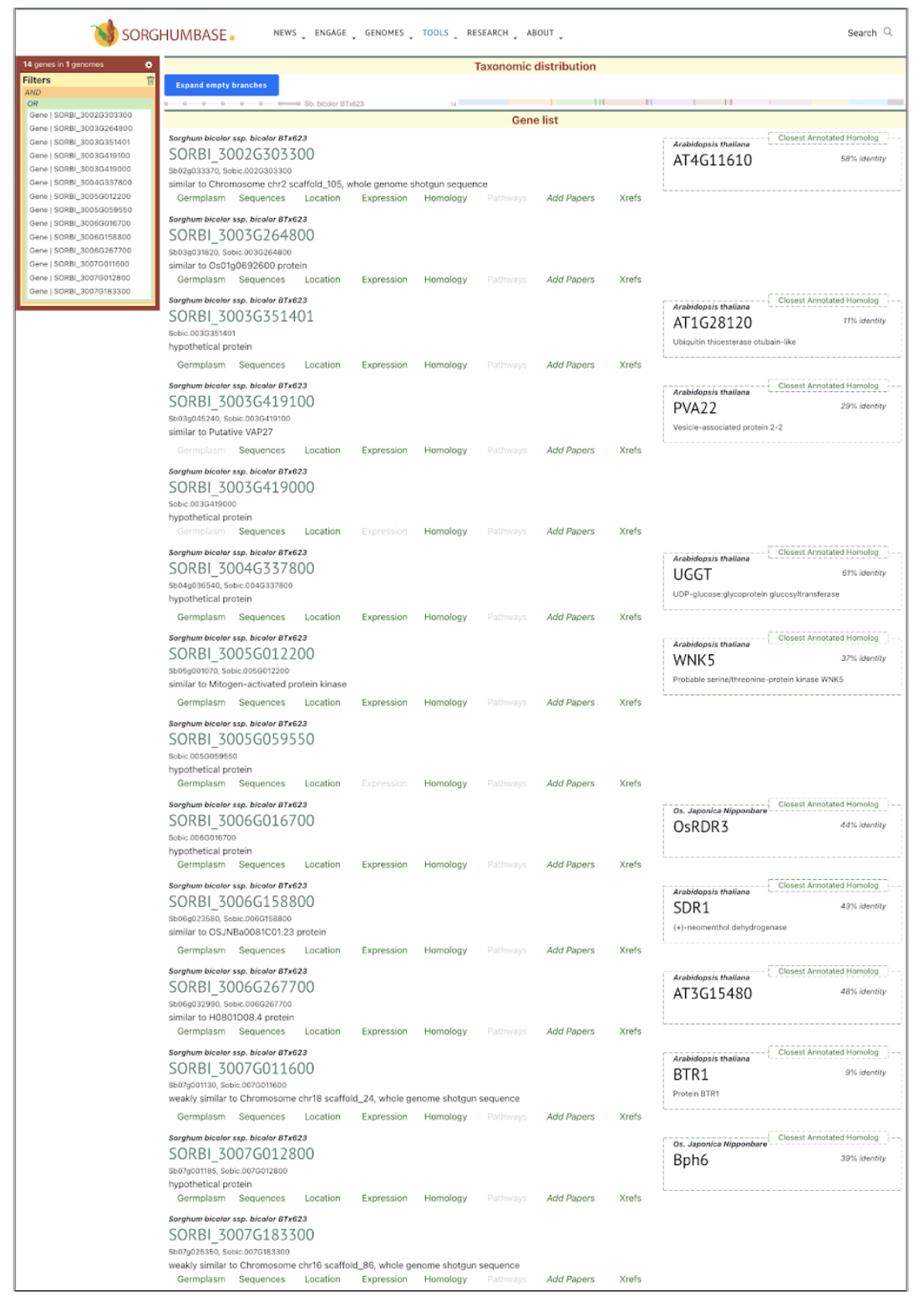
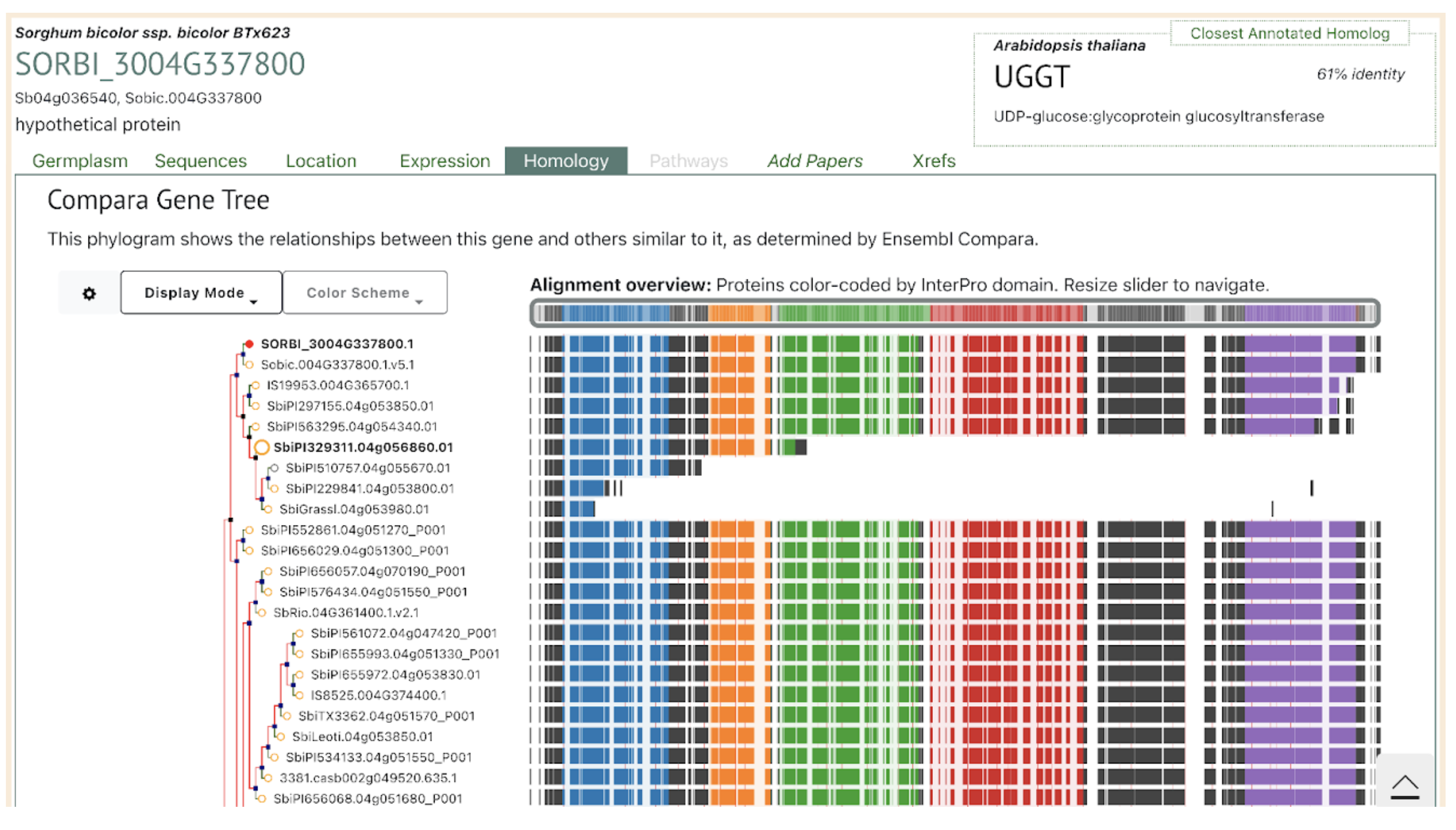
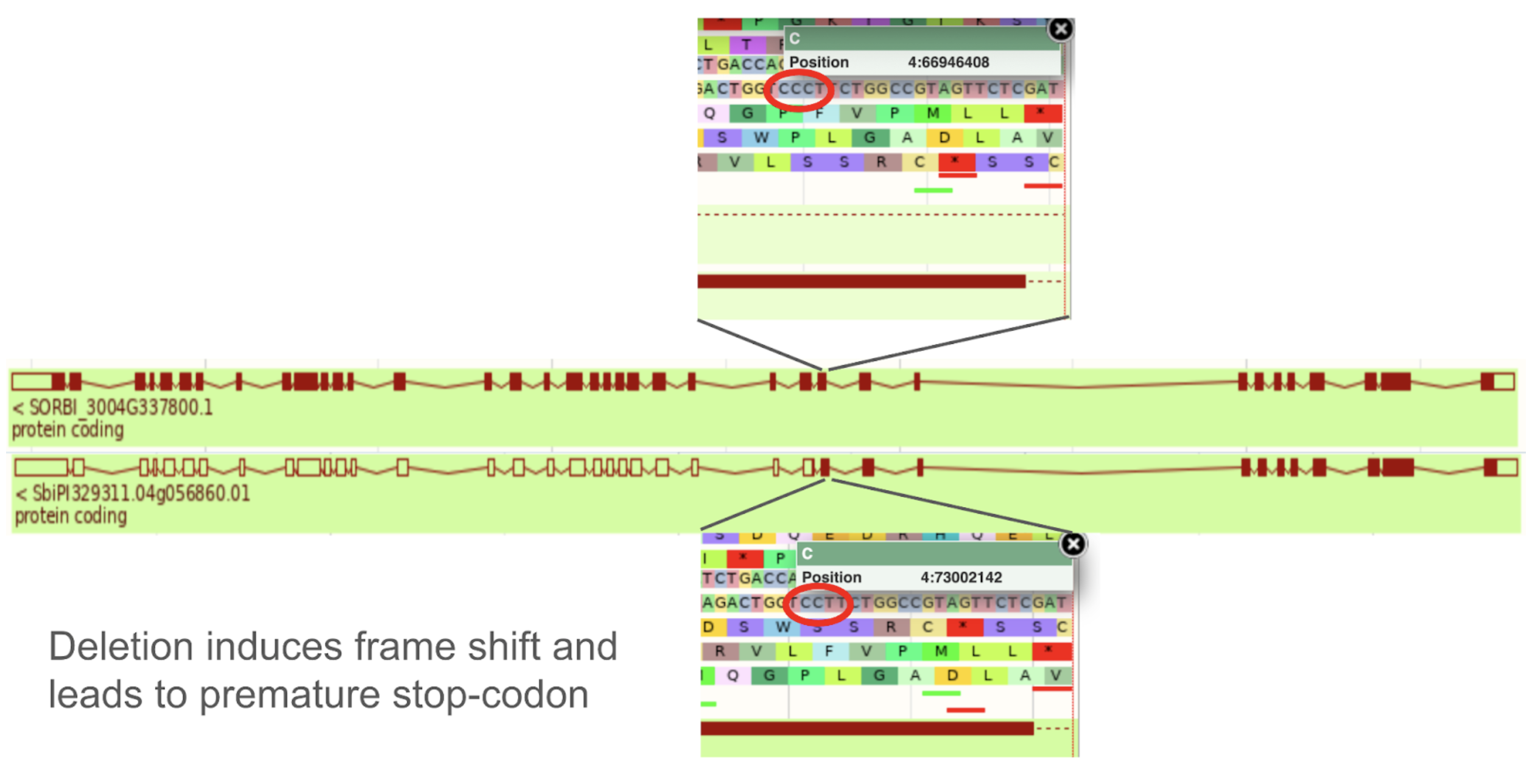
Reference:
Tsehaye Y, Menamo TM, Demelash H, Abay F, Tadesse T, Bantte K. Multi-locus genome-wide association study reveals genomic regions underlying root system architecture traits early growth stage of sorghum germplasm. Sci Rep. 2025 Aug 28;15(1):31727. PMID: 40877421. doi: 10.1038/s41598-025-99012-w. Read more

