SbWRKY51, a group II WRKY transcription factor in sorghum, enhances salt tolerance by maintaining ion and ROS homeostasis and promoting lignin biosynthesis to strengthen cell wall integrity under salt stress.
Keywords: Lignin, ROS, Salt stress, Sorghum, WRKY
Sorghum is recognized for its strong tolerance to salinity, making it a valuable system for elucidating the molecular mechanisms of salt tolerance and identifying candidate genes for crop improvement. Scientists from Shandong Normal University identified a novel WRKY transcription factor, SbWRKY51, characterized it as a positive regulator of salt stress response in sorghum. Sequence and phylogenetic analyses revealed that SbWRKY51 belongs to the group II WRKY family and shares high similarity with AtWRKY11 from Arabidopsis thaliana. SbWRKY51 localized to the nucleus and exhibited transcriptional activation activity, suggesting a role in gene regulation. Transgenic Arabidopsis and sorghum lines overexpressing SbWRKY51 showed enhanced tolerance to salt stress, as evidenced by higher germination rates, greater root length, increased biomass, and improved maintenance of Na⁺/K⁺ homeostasis compared to wild-type and mutant lines. These results indicate that SbWRKY51 enhances salt tolerance by modulating ion balance and stress-related physiological processes.
Further analyses revealed that SbWRKY51 contributes to oxidative stress regulation and cell wall modification under salt stress. SbWRKY51-overexpressed plants accumulated less reactive oxygen species (ROS) and exhibited higher activities of antioxidant enzymes such as catalase (CAT) and superoxide dismutase (SOD). Correspondingly, ROS-scavenging genes (e.g., APX, CAT, GPX6) were upregulated, while the ROS-producing gene RBOHA was downregulated. Gene Ontology and KEGG enrichment analyses indicated that SbWRKY51 target genes were enriched in oxidative stress response and phenylpropanoid biosynthesis pathways. Consistent with this, SbWRKY51-overexpressed sorghum lines showed elevated expression of lignin biosynthesis genes (SbPAL, SbC4H, SbCOMT, etc.) and higher lignin accumulation, which may enhance structural integrity and limit Na⁺ influx. Collectively, these findings demonstrate that SbWRKY51 improves salt tolerance in sorghum by coordinating ROS homeostasis and lignin biosynthesis.
SorghumBase examples:
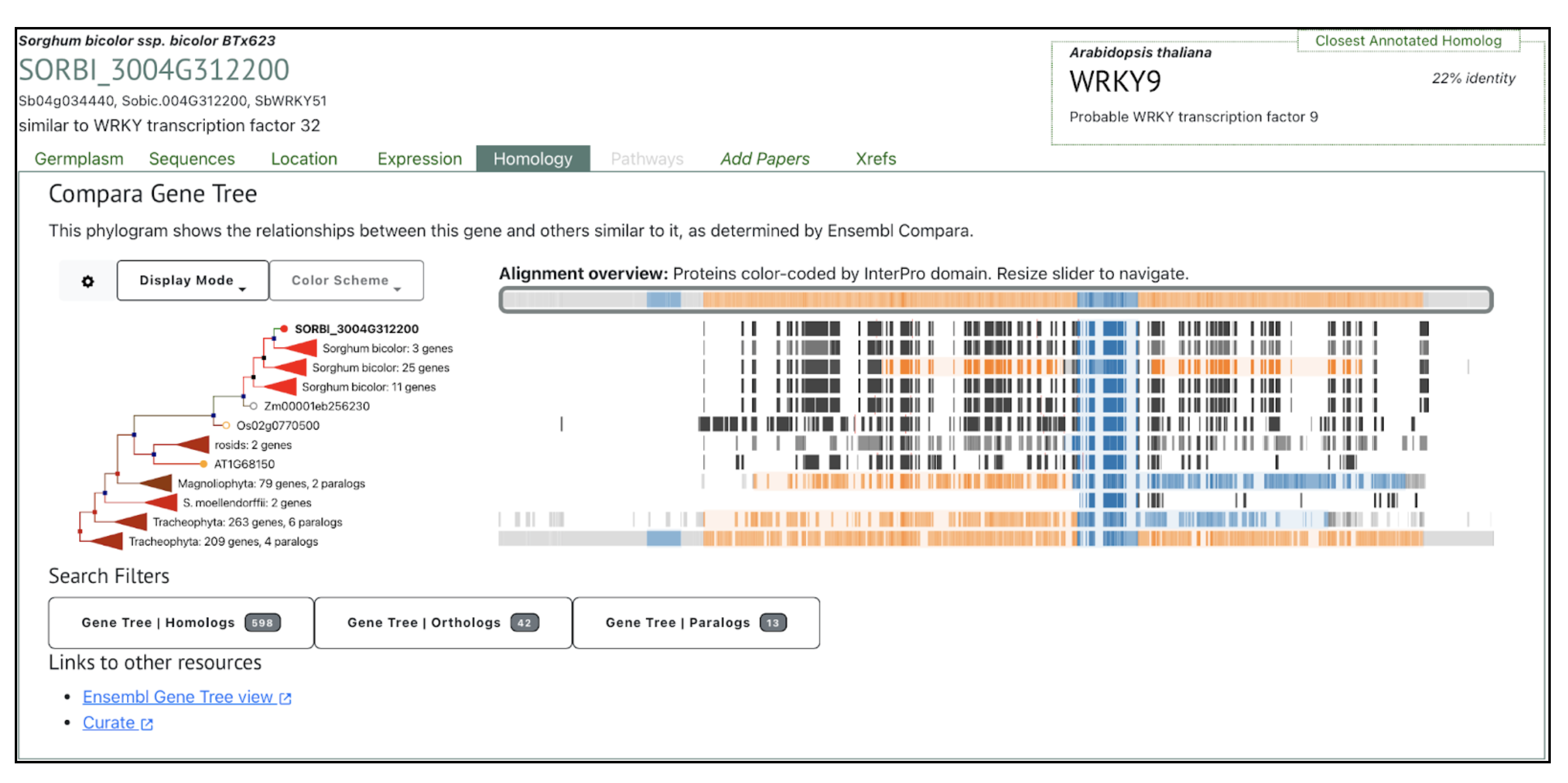
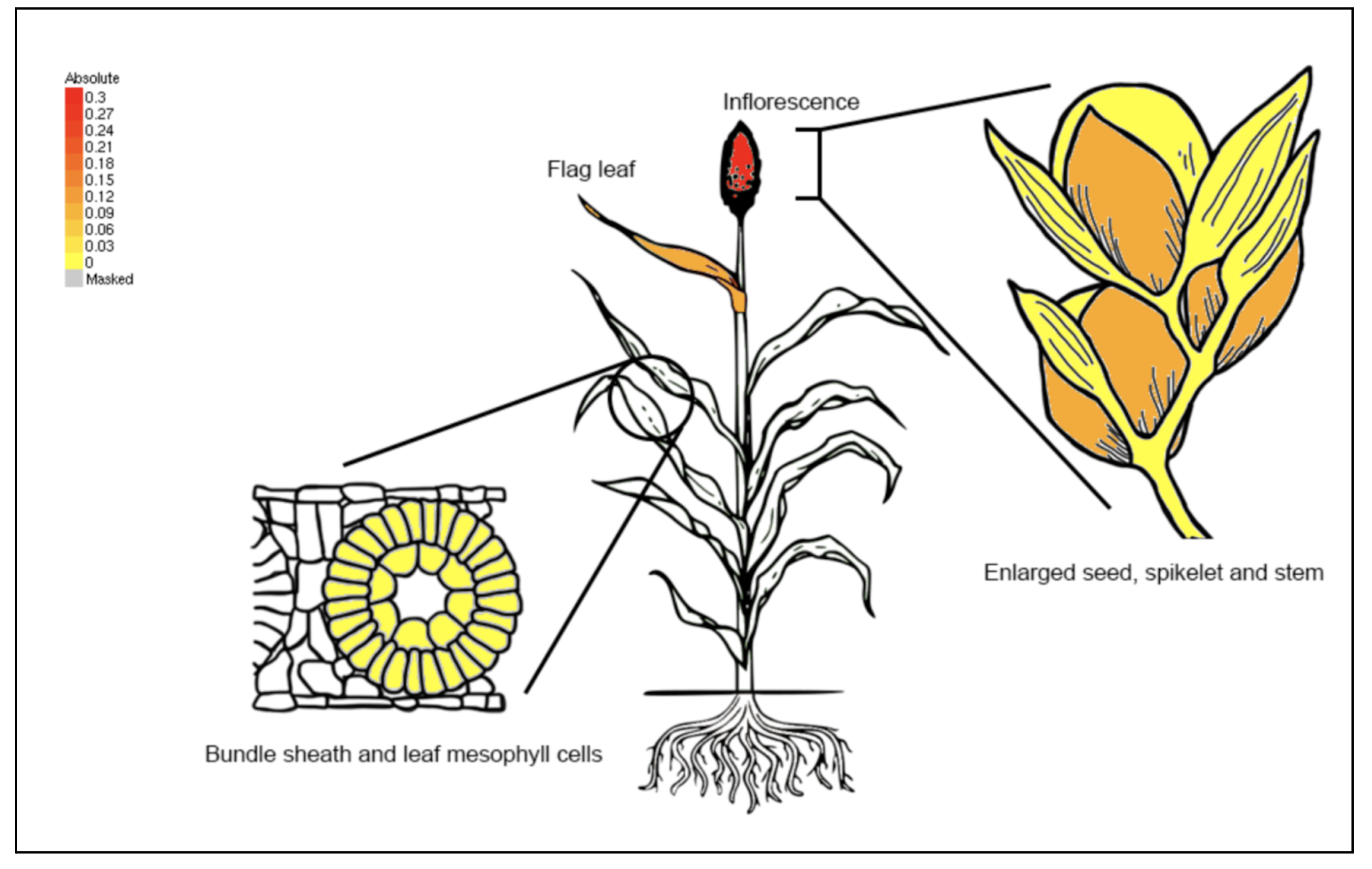
Reference:
Wang X, Li H, Wang J, Song J, Sui N. The WRKY Transcription Factor SbWRKY51 Positively Regulates Salt Tolerance of Sorghum. Plant Sci. 2025 Sep 2;362:112741. PMID: 40907827. doi: 10.1016/j.plantsci.2025.112741. Read more