Linalool enhances sorghum resistance to Colletotrichum sublineola-induced anthracnose by activating salicylic acid–mediated defense signaling and directly inhibiting fungal growth through membrane disruption and oxidative stress.
Keywords: anthracnose, antifungal mechanism, linalool, plant defense, sorghum, terpene synthase, transcriptomic analysis
Anthracnose, caused by Colletotrichum sublineola, severely limits sorghum yield by inducing premature leaf necrosis and reducing photosynthetic capacity, with losses exceeding 50% under epidemic conditions. Understanding the molecular basis of resistance is therefore essential for sustainable disease management. Transcriptomic and biochemical analyses have revealed that secondary metabolites, particularly terpenoids, play central roles in defense activation following infection. Researchers from Qingdao Agricultural University and Yangzhou University identified a strong correlation between linalool accumulation and anthracnose resistance, suggesting that this monoterpene contributes to pathogen suppression and immune priming. Unlike jasmonic acid (JA)-related defense, linalool-induced responses appear to operate through salicylic acid (SA)-mediated signaling. Linalool pretreatment reduced lesion formation and mitigated declines in photosynthetic and physiological parameters under infection, highlighting its dual role as both a signaling molecule and an antimicrobial compound.
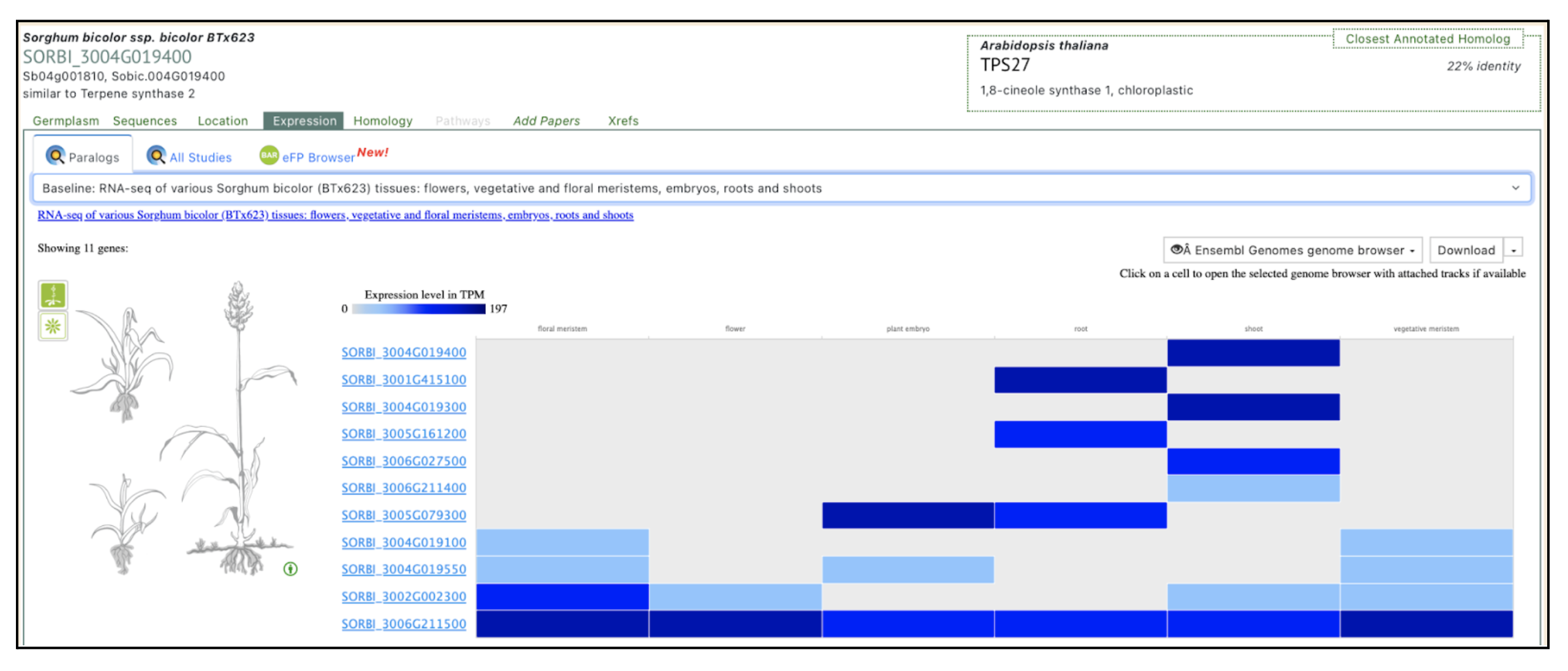
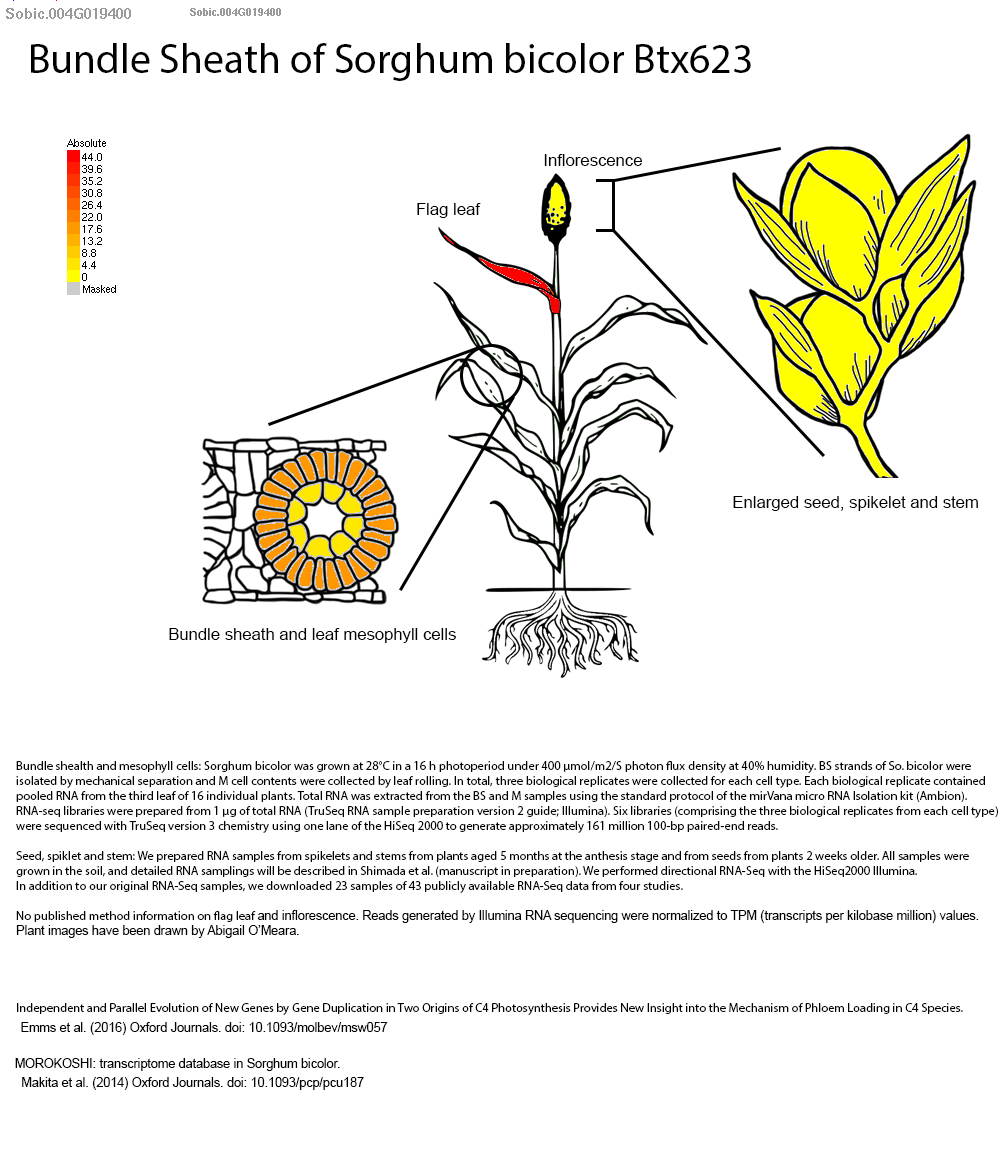
Mechanistic assays demonstrated that linalool exerts direct antifungal effects against C. sublineola by disrupting cell membranes, inhibiting spore germination, and inducing oxidative stress through lipid peroxidation and reactive oxygen species accumulation. Xiong et al. further identified Sobic.004G019400 as a linalool synthase gene whose expression is upregulated in resistant genotypes and likely regulated by defense-associated transcription factors (bHLH, WRKY, MYB, and NAC). These findings underscore the potential of enhancing linalool biosynthesis—via genetic engineering or exogenous application—as an eco-friendly alternative to chemical fungicides. Future dual RNA-seq analyses are proposed to map the full sorghum–pathogen interaction landscape, advancing integrated pest management strategies and guiding the breeding of anthracnose-resistant sorghum cultivars.
SorghumBase Examples:
Authors have identified Sobic.004G019400 as the key terpene synthase (TPS) gene responsive to fungal infection through transcriptome profiling. This gene has been selected by authors as a molecular target for breeding anthracnose-resistant sorghum varieties. We selected this gene to explore more on sorghumbase (https://sorghumbase.org/).
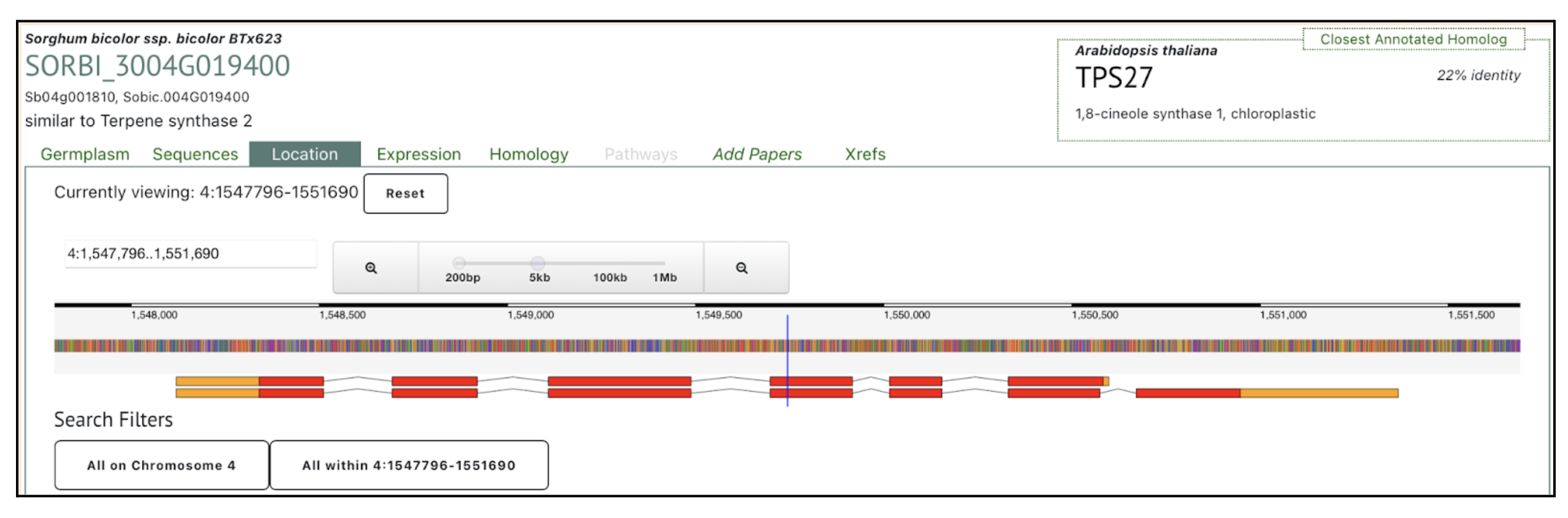
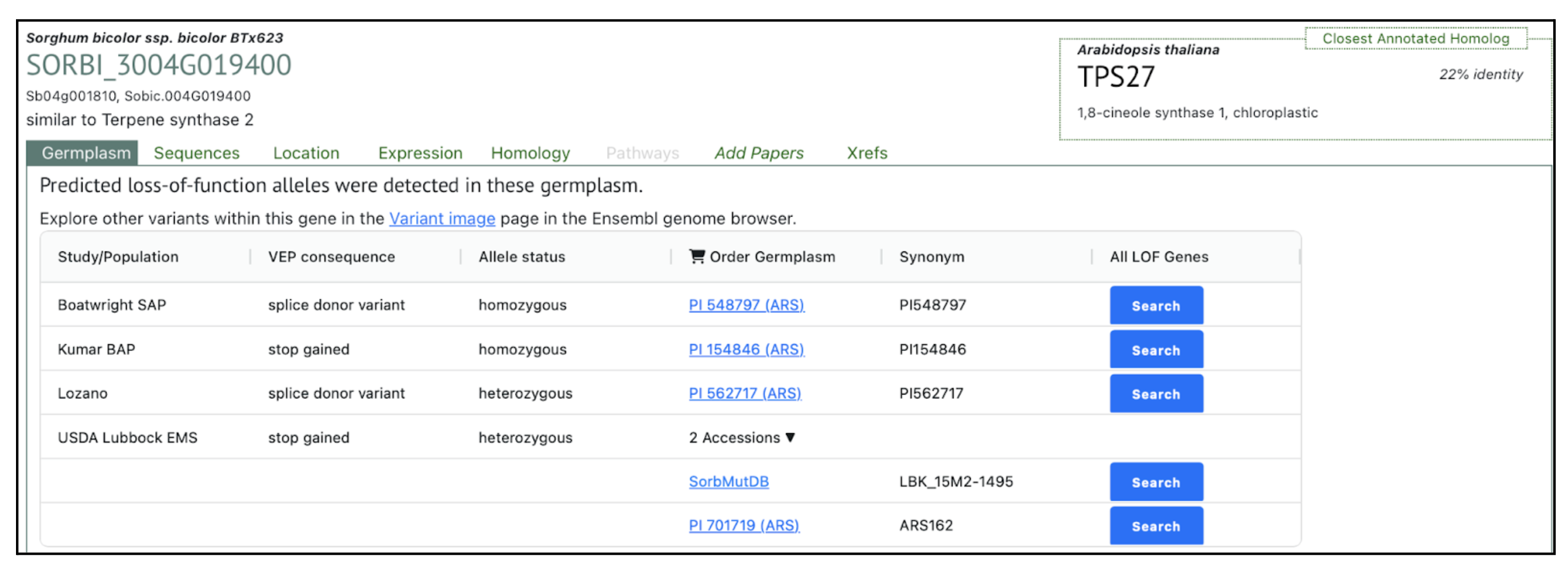
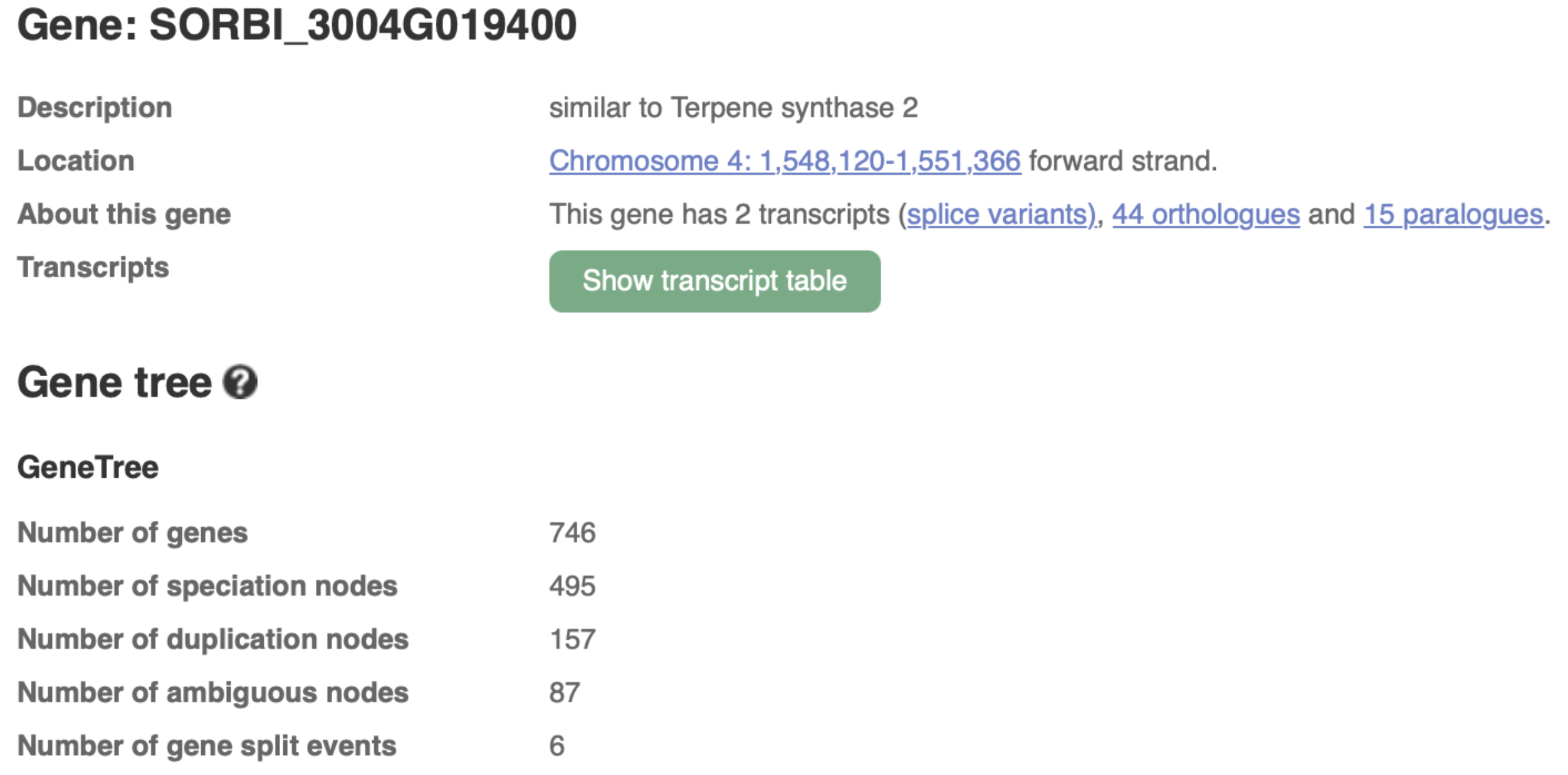
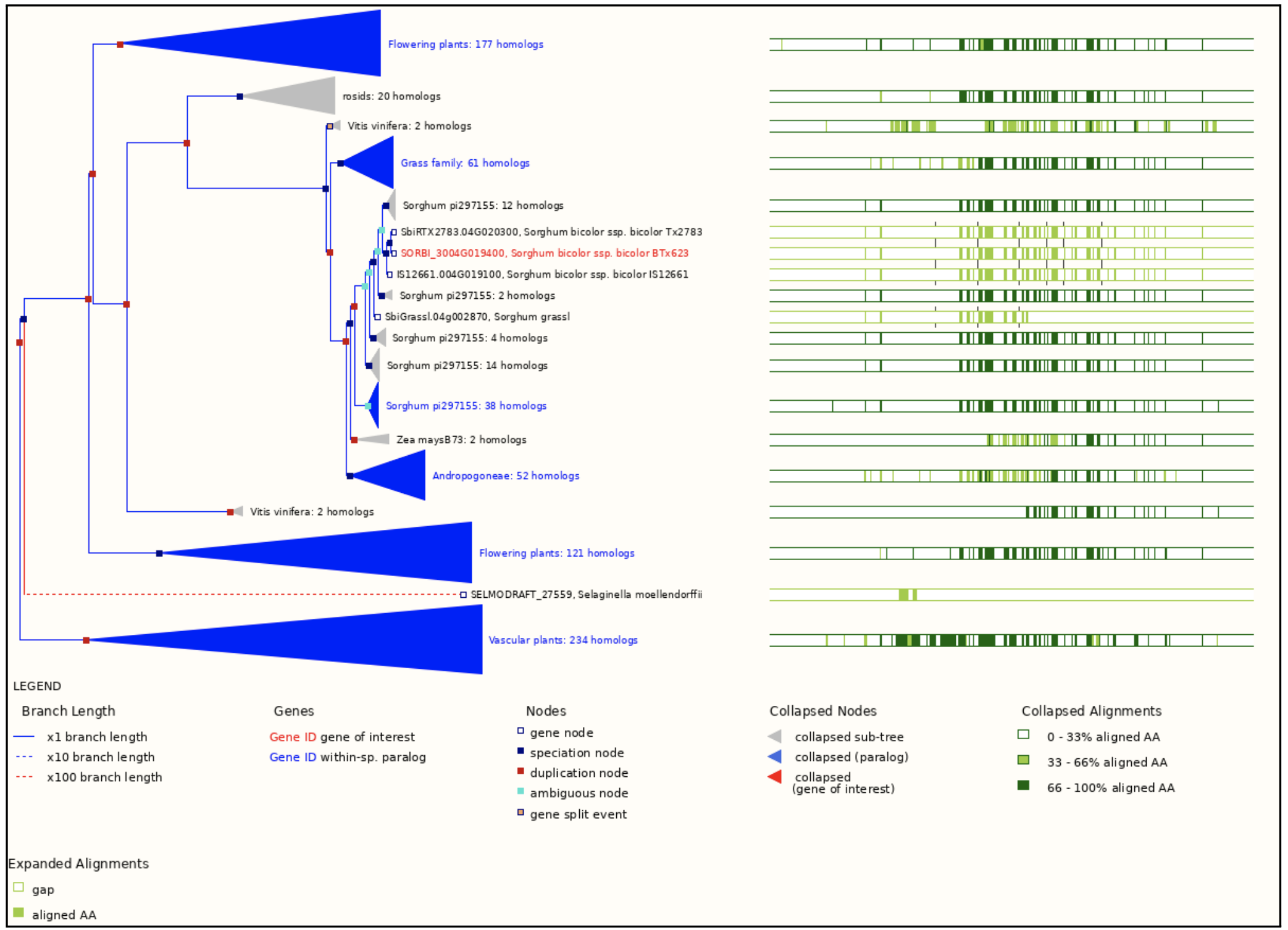
Xiong W, Jia X, Wang T, Wei G, Guo Y. Unveiling the mechanisms for plant-derived linalool control of anthracnose in sorghum. Pest Manag Sci. 2025 Sep 18. PMID: 40964958. doi: 10.1002/ps.70240. Read more