Sorghum polyamine oxidase genes exhibit distinct structural, evolutionary, and regulatory specializations that shape tissue- and genotype-specific stress responses, with SbPAO5 and SbPAO6 emerging as key contributors to drought tolerance and promising targets for crop improvement.
Keywords: Gene expression, Genome-wide, Polyamine oxidase, Sorghum, promoters
Polyamine oxidases are central players in polyamine catabolism and plant stress adaptation, yet their functional landscape in sorghum has remained largely unexplored. Building on our earlier sequence-based identification of sorghum PAOs (Ebeed et al., 2025), this study delivers the first comprehensive evolutionary, structural, and regulatory characterization of the SbPAO gene family. My findings reveal distinct duplication patterns, subcellular targeting signals, and stress-responsive regulatory elements, highlighting the specialized roles of SbPAO4–6, particularly under drought conditions. The strong induction of SbPAO5 and SbPAO6 in the tolerant genotype Dorado, alongside conserved catalytic features and subfunctionalized protein folds, underscores their potential as promising candidates for molecular breeding. I believe these insights will accelerate functional validation and create new opportunities to enhance sorghum resilience through targeted genetic improvement – Ebeed
Polyamine oxidases (PAOs) play essential roles in polyamine catabolism and are key contributors to plant growth, development, and stress adaptation. Researcher from Damietta University and Academy of Scientific Research and Technology (ASRT) conducted comprehensive characterization of the sorghum PAO (SbPAO) gene family which revealed six members exhibiting distinct structural, evolutionary, and regulatory features. Motif analysis identified ten conserved motifs, including a putative novel motif present in all SbPAOs, and multiple sequence alignment highlighted residues essential for FAD binding and amine oxidation. Predicted protein structures showed conserved catalytic cores alongside specialized folds in SbPAO5 and SbPAO6, consistent with subfunctionalization. Subcellular localization predictions further suggested divergent biochemical roles, with SbPAO4 and SbPAO5 harboring conserved peroxisomal targeting signals, implicating these proteins in compartmentalized polyamine turnover and localized H₂O₂ signaling. Phylogenetic reconstruction placed SbPAOs into four distinct clades together with PAOs from Arabidopsis, rice, and maize, supporting both shared ancestry and functional divergence. Synteny and chromosomal mapping identified segmental (SbPAO1/2) and tandem (SbPAO3/4/5) duplications, indicating gene family expansion typical of stress-associated pathways.
Integration of co-expression networks, promoter motif analysis, and stress-responsive transcript profiling revealed that SbPAO genes participate broadly in abiotic stress regulation. Promoters were enriched in stress-, hormone-, and light-responsive cis-elements, while protein–protein interaction networks linked SbPAOs to polyamine metabolism and defense signaling. Under drought, the tolerant genotype Dorado displayed strong induction of SbPAO4–6, coordinated polyamine accumulation, and stable grain weight, whereas the sensitive genotype Giza 15 showed limited PAO activation and reduced yield. The pronounced drought-responsive behavior, specialized structures, and regulatory connectivity of SbPAO5 and SbPAO6 position them as promising targets for genetic improvement. Overall, these findings demonstrate the evolutionary diversification and functional specialization of PAO genes in sorghum and highlight their potential for enhancing drought resilience through molecular breeding or biotechnological strategies.
SorghumBase Examples:
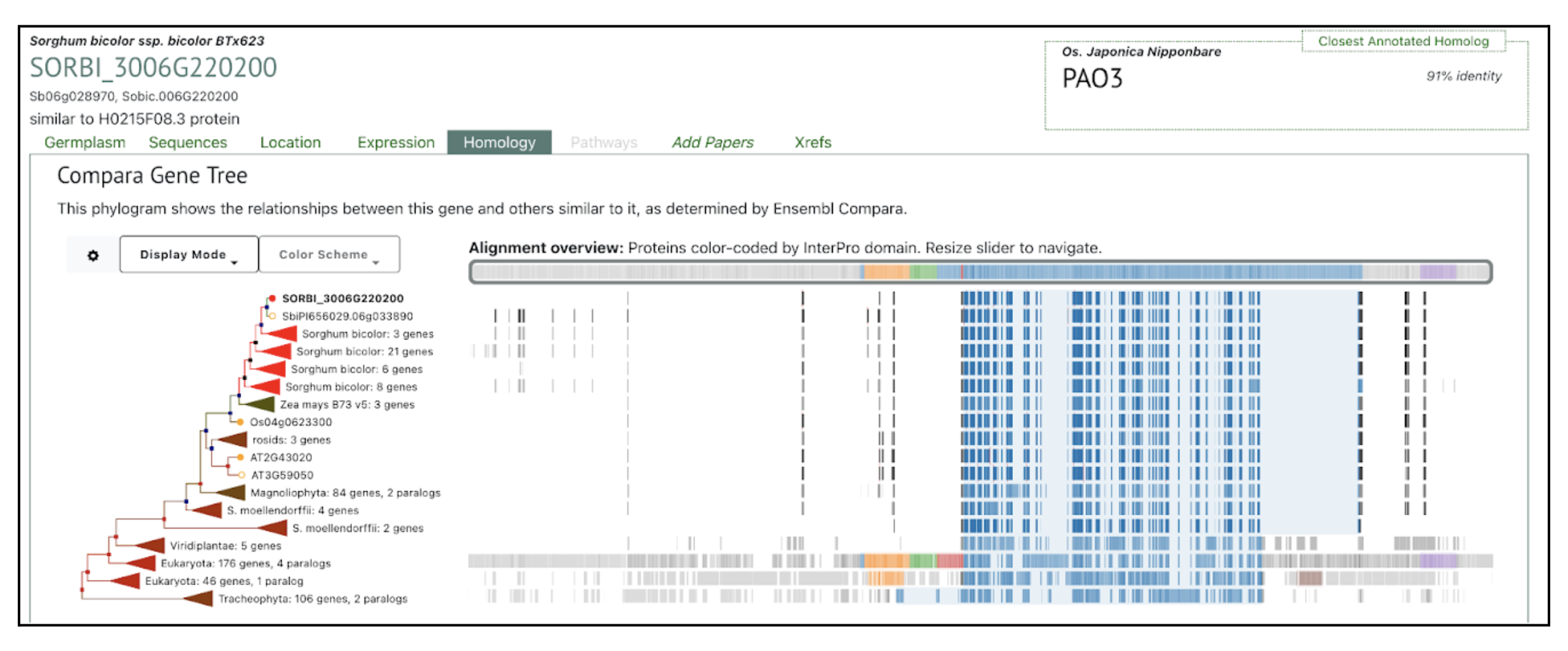
The SorghumBase Homology panel positions SbPAO5 within a conserved monocot clade of polyamine oxidase genes, alongside multiple Sorghum bicolor paralogs and close orthologs from maize and rice. The closest annotated homolog, rice PAO3 (91% identity) further underscores deep conservation across cereals. The alignment overview to the right shows that SbPAO5 contains the IPR002937 Amino_oxidase (Amine oxidase) domain, a catalytic domain shared by 457 of 472 genes (96.8%) in this gene tree. This high degree of domain conservation matches the study’s comparative genomics findings, which indicate that the PAO gene family underwent segmental duplication while retaining the core amine-oxidase catalytic machinery essential for polyamine turnover and drought-responsive signaling.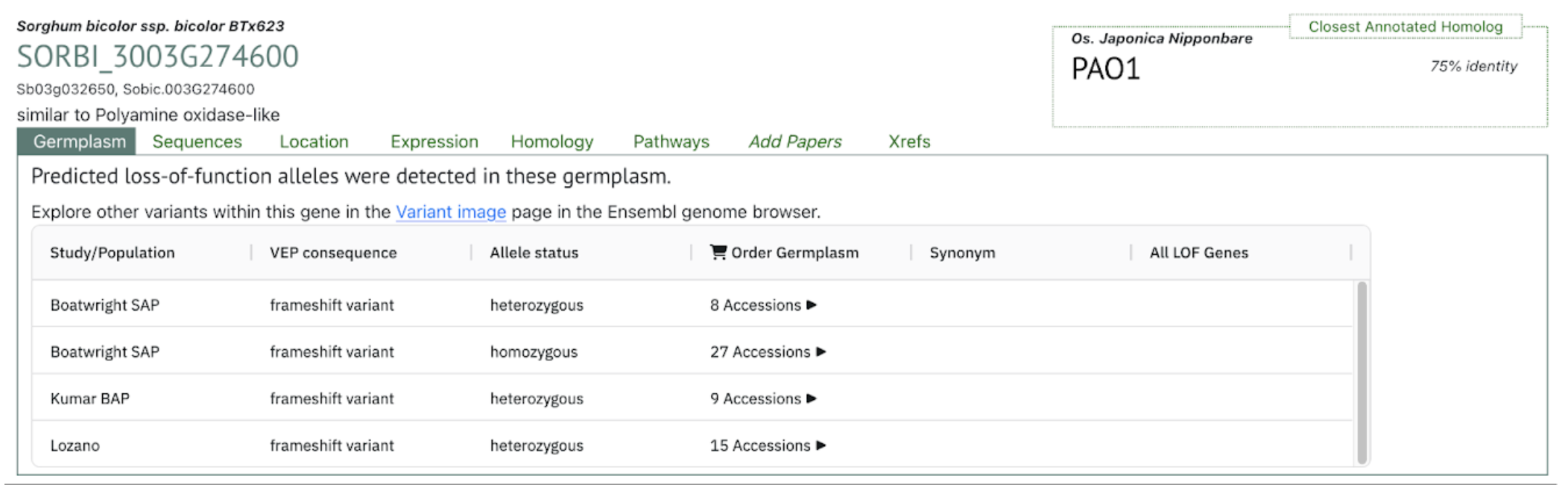
The SorghumBase Germplasm view shows that SbPAO6 carries frameshift loss-of-function (LOF) variants in several diversity panels, including the Boatwright SAP, Kumar BAP, and the Lozano collection. These variants occur in both heterozygous and homozygous states, with up to 27 accessions carrying homozygous frameshift alleles in the Boatwright SAP panel. This indicates that natural LOF variants of SbPAO6 are present and moderately frequent within global sorghum diversity, providing valuable genetic resources for testing the functional contribution of SbPAO6 to drought tolerance and polyamine-oxidase–mediated stress responses.
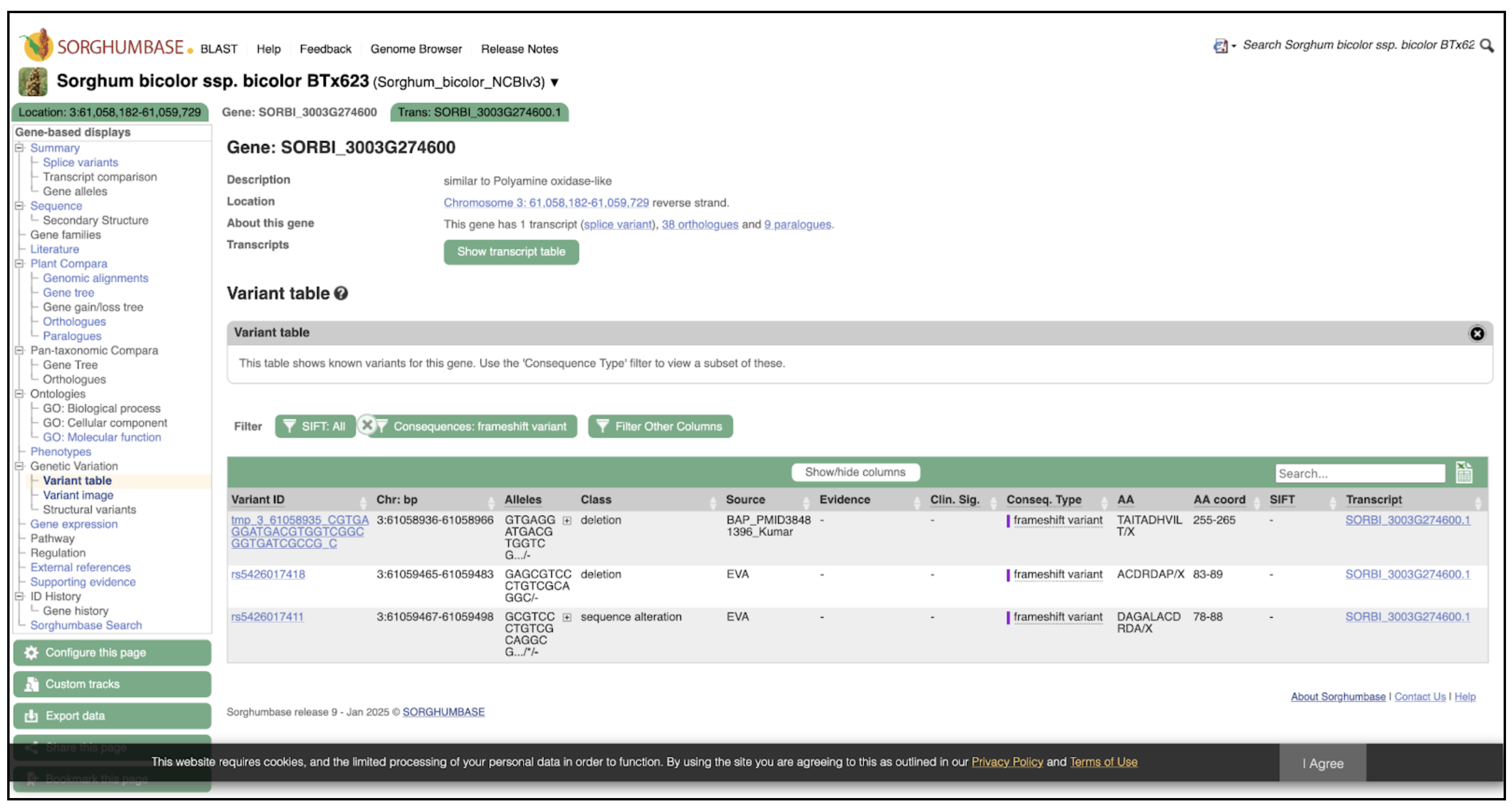
The Variant Table lists three independent frameshift variants in SbPAO6, each classified as a deletion or sequence alteration predicted to disrupt the protein-coding sequence. These variants originate from curated population studies, including the Kumar BAP panel and EVA submissions. Amino acid consequence annotations indicate that frameshifts occur at positions 78–89 and 255–265, likely abolishing or severely reducing PAO catalytic activity. Together with the germplasm-level summaries shown in Figure 2A, these entries demonstrate the presence of naturally occurring functional knockouts of SbPAO6, offering strong candidates for genotype–phenotype association studies and targeted functional validation.
Reference:
Ebeed HT. Comparative genomics and expression analysis of polyamine oxidase gene family in Sorghum bicolor reveals functional specialization, gene duplication, and role in drought resilience. BMC Genomics. 2025 Oct 28;26(1):966. PMID: 41152702. doi: 10.1186/s12864-025-12125-4. Read more
Related Project Websites:
- Heba T. Ebeed’s ORCID profile: https://orcid.org/0000-0002-3928-8123
- Heba T. Ebeed’s Web of Science Author Profile: https://www.webofscience.com/wos/author/record/R-1477-2016