Liu et al. found that though alkali stress disrupts growth, osmotic balance, and cellular stability in sorghum, the tolerant genotype Z14 counters these effects through stronger antioxidant defenses, enhanced osmotic regulation and rapid activation of stress-responsive genes and signaling pathways.
Keywords: Sorghum bicolor (L.), alkali stress, antioxidant activity, signal transduction, transcriptome
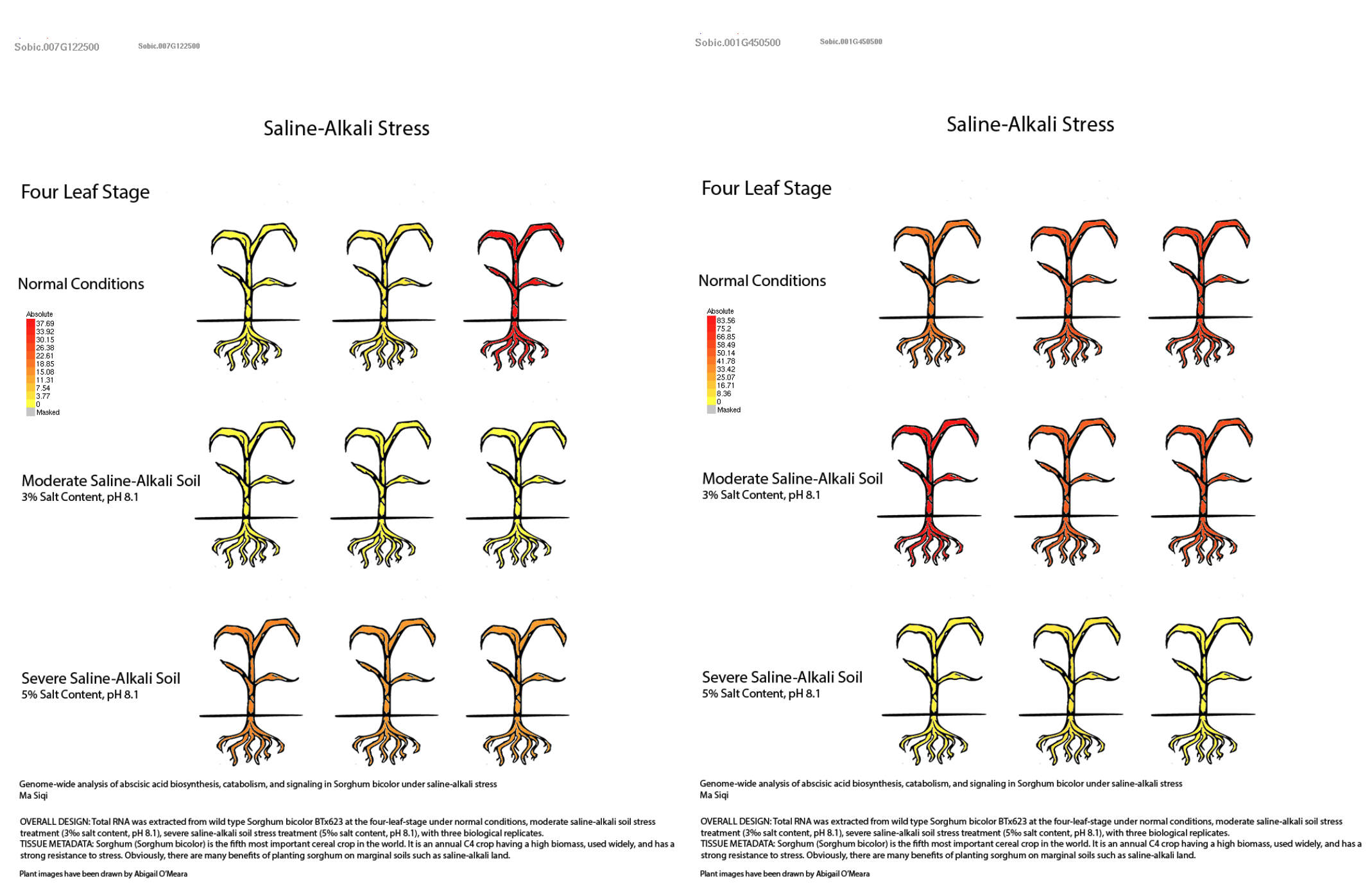
Alkali stress is an expanding global constraint on crop productivity, arising from factors such as reduced rainfall, excessive fertilization, improper irrigation, and industrial pollution. As alkalinity intensifies, plants must reestablish osmotic balance and ion homeostasis to avoid growth inhibition and cell damage. In this study, conducted by scientists from Heilongjiang Bayi Agricultural University and collaborating institutions, plant height and root elongation in sorghum was severely suppressed under alkali stress (80 mM NaHCO₃) in both tolerant (Z14) and sensitive (Z1) genotypes, with more pronounced inhibition in Z1. Elevated levels of reactive oxygen species (ROS) and malondialdehyde (MDA) indicated oxidative damage, particularly in Z1, whereas Z14 showed stronger accumulation of osmotic regulators—especially soluble proteins—and higher activities of antioxidant enzymes such as SOD, POD, CAT, and APX. These physiological traits suggest that Z14 more effectively restricts ROS accumulation and maintains cellular stability under stress. Transcriptomic analyses revealed that Z14 rapidly activates stress-responsive genes within the first 12 hours of alkali exposure, supporting early molecular adaptation.
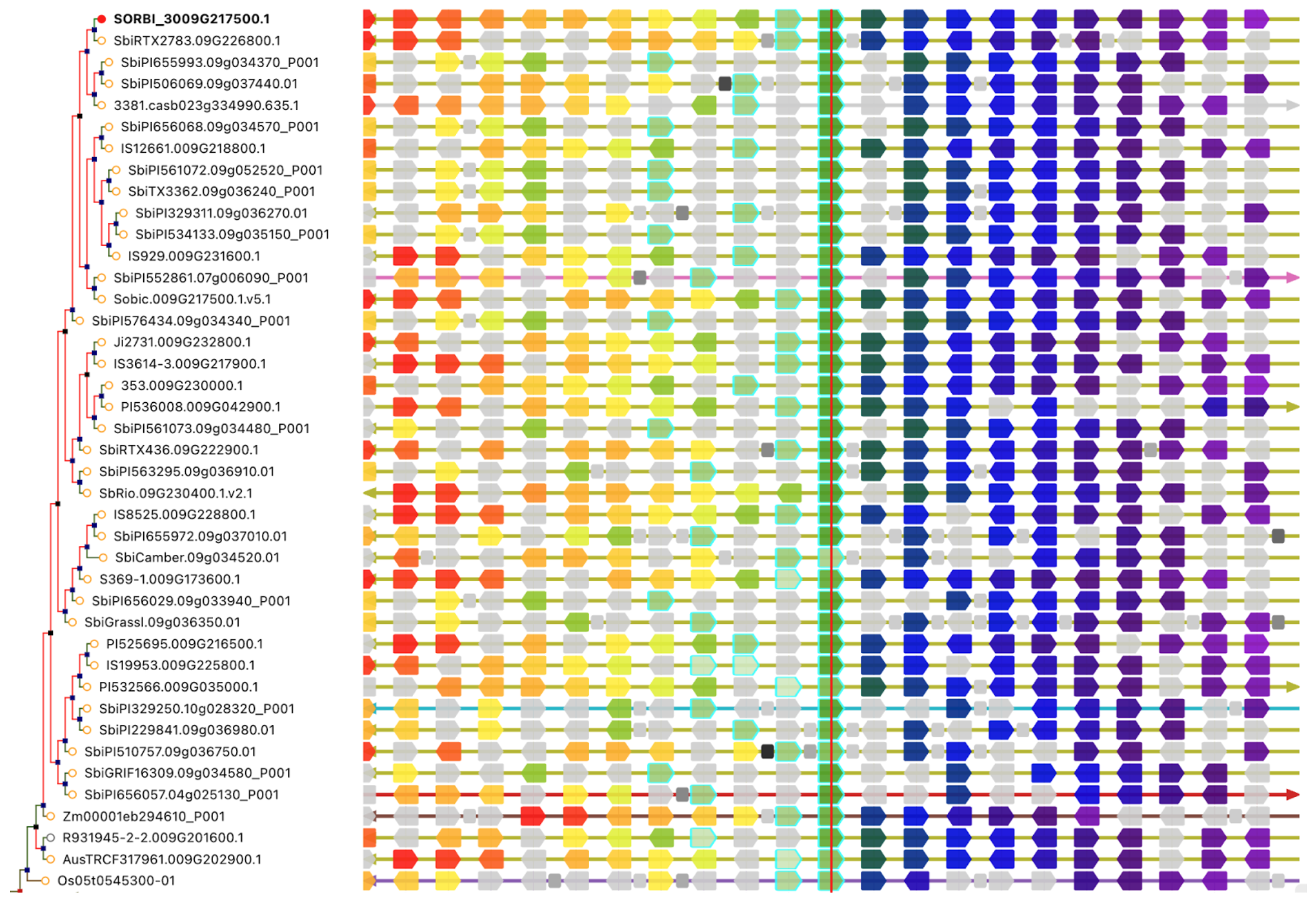
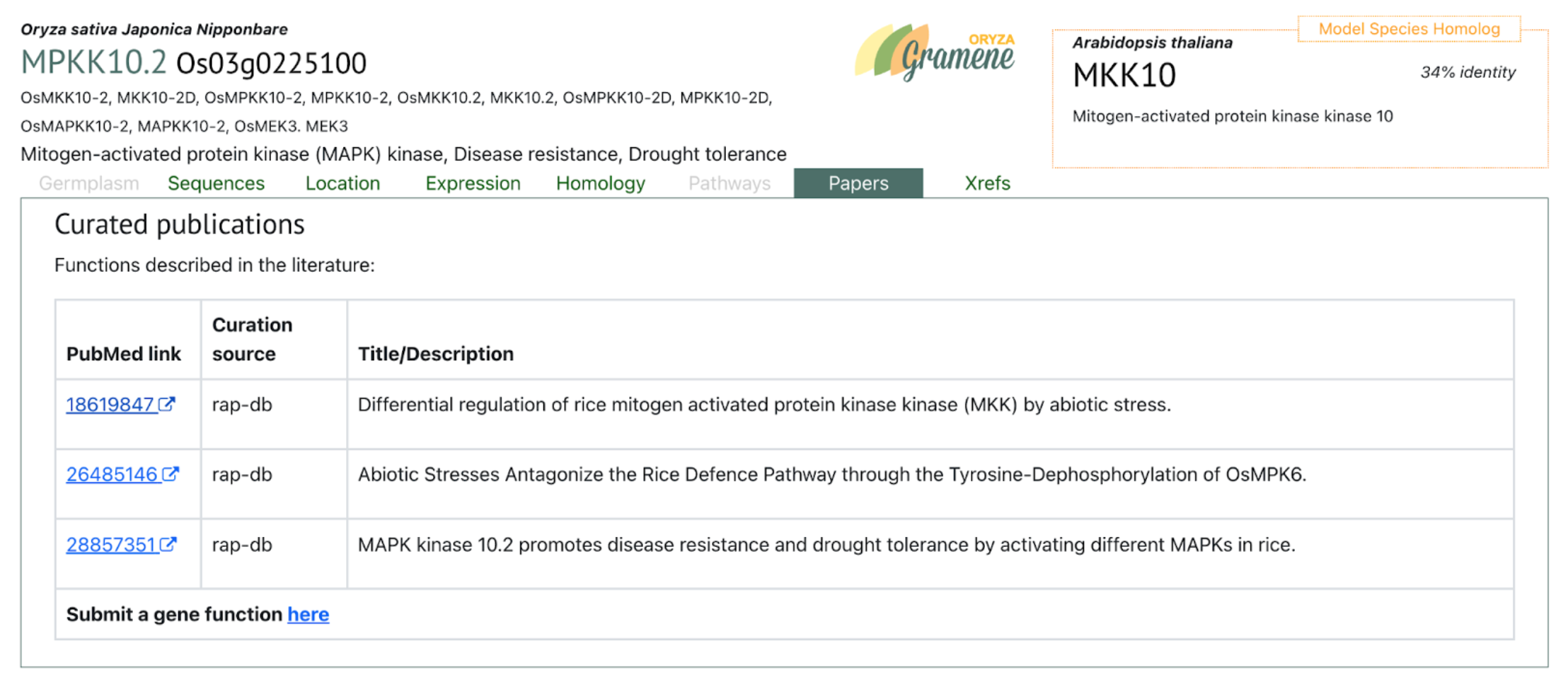
Large transcription factor families—including bHLH, ERF, MYB, and MYB-related proteins—were strongly induced in Z14, consistent with their known roles in controlling downstream stress-response pathways related to ion transport, osmotic adjustment, and antioxidant defense. Key signal transduction components were also activated, including ABA-related PP2Cs, MAPK cascade genes, and Ca²⁺-dependent signaling elements (CPKs, CIPKs, CBLs). Additionally, genes involved in maintaining ion homeostasis (e.g., SOS, NHX, HKT, and HAK family members) were upregulated, suggesting coordinated regulation of Na⁺/H⁺ balance under high pH conditions. Together, these responses form an integrated adaptive network in sorghum seedlings, enabling the maintenance of photosynthetic capacity, osmotic stability, ROS detoxification, and membrane integrity during alkali stress.
SorghumBase Examples:
Reference:
Liu X, Wang B, Zhao Y, Chu M, Yu H, Gao D, Wang J, Li Z, Liu S, Li Y, Wei Y, Wei J, Xu J. Physiological and Transcriptional Responses of Sorghum Seedlings Under Alkali Stress. Plants (Basel). 2025 Oct 9;14(19):3106. PMID: 41095246. doi: 10.3390/plants14193106. Read more