Genetic modeling and high-density QTL mapping reveal that sorghum plant height and brix content are governed by interacting major genes and polygenes, share co-localized loci that explain their phenotypic correlation, and are influenced by auxin- and carbon-fixation–related candidate genes that offer targets for breeding improved varieties.
Keywords: Sorghum, Plant height, Brix content, Genetic linkage map, QTL mapping
Cultivated sorghum exhibits substantial phenotypic diversity, with plant height and brix content representing key agronomic traits for biomass production and sugar yield. Scientists from Northwest A&F University and Liaoning Academy of Agricultural Sciences used mixed inheritance models and multi-environment phenotyping to dissect the genetic architecture underlying these traits. Both plant height and brix content displayed near-normal phenotypic distributions, consistent with control by major genes and polygenes. Genetic model analyses revealed that plant height is regulated by two major gene pairs with epistatic interactions in addition to additive polygenes, while brix content is influenced by three major gene pairs alongside polygenic effects. Major genes contributed more strongly to heritability than polygenes, indicating that early-generation selection is effective for both traits. A high-density sorghum linkage map (1129.97 cM, 0.55 cM average interval) enabled the identification of QTL across chromosomes 1, 5, 6, 7, and 8, including loci that overlapped with previously reported QTL for plant height and stalk sugar concentration. Co-localized QTL for plant height and brix content, particularly on chromosome 7, support the observed phenotypic correlation between the traits, suggesting either tight linkage or pleiotropy.
Candidate gene analysis within the shared QTL intervals identified genes associated with auxin biosynthesis and transport—including homologs of YUCCA8 and ABC transporter family proteins—as likely regulators of plant height. Similarly, three genes encoding malate dehydrogenase, key components of the C₄ carbon-fixation pathway, were identified as strong candidates influencing brix content. These findings provide new insights into the genetic regulation of height–sugar relationships in sorghum and offer molecular targets for marker-assisted breeding. By integrating genetic models, high-resolution QTL mapping, and candidate gene analyses, this study advances the mechanistic understanding of quantitative trait variation and supports efforts to develop sorghum varieties with improved biomass yield and sugar accumulation.
SorghumBase Examples:
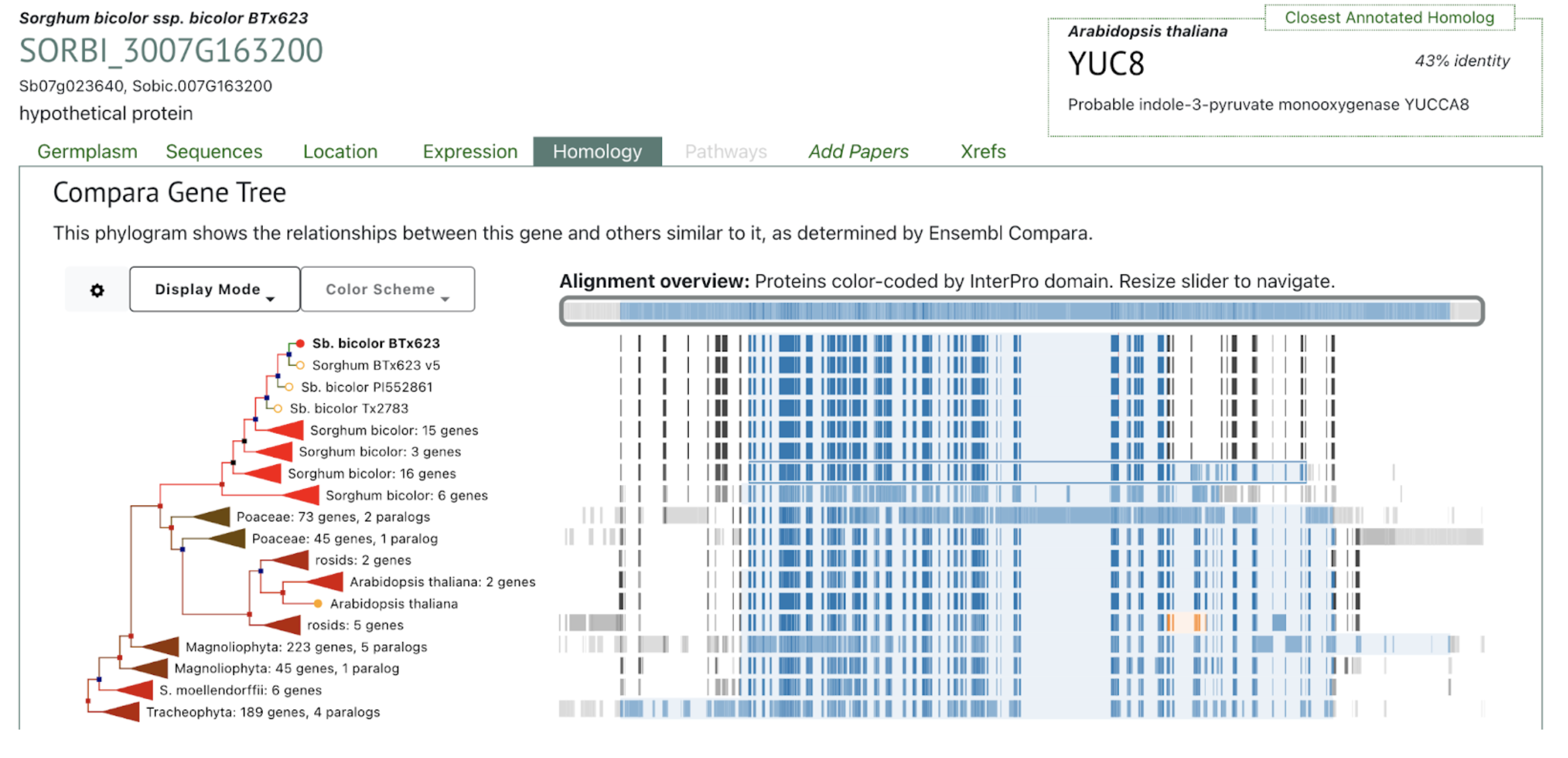
The phylogram places Sobic.007G163200 within a conserved YUCCA-like monooxygenase clade, grouping with several Sorghum bicolor paralogs and orthologous genes across grasses and dicots. The closest annotated homolog, Arabidopsis YUC8 (43% identity), reflects shared roles in indole-3-pyruvate–dependent auxin biosynthesis. The alignment overview highlights strong conservation of diagnostic YUCCA domains across species, consistent with the gene’s proposed involvement in regulating internode elongation and height variation in sorghum.
The SorghumBase Compara Gene Tree places Sobic.007G166200 within a large, deeply conserved malate dehydrogenase (MDH) clade spanning grasses, dicots, and basal plant lineages. The closest annotated homolog, Arabidopsis MDH1 (30% identity), confirms conservation of core enzymatic domains involved in NAD(P)-dependent malate oxidation. The accompanying neighborhood-conservation panel illustrates strong synteny: genes flanking Sobic.007G166200 share conserved order and family identity across multiple species, emphasizing the stability of this metabolic locus. Together, these patterns support the gene’s proposed function in carbon fixation and sugar accumulation, consistent with its role as a brix QTL candidate in the study.
Shi X, Duan Y, Chai S, Guo Y, Guo S, Wang C, Li S, Liu D, Feng B, Lu F, Yang P. QTL mapping and candidate genes prediction for plant height and brix content in sorghum [Sorghum bicolor (L.) Moench]. Theor Appl Genet. 2025 Sep 3;138(9):238. PMID: 40900327. doi: 10.1007/s00122-025-05024-5. Read more
