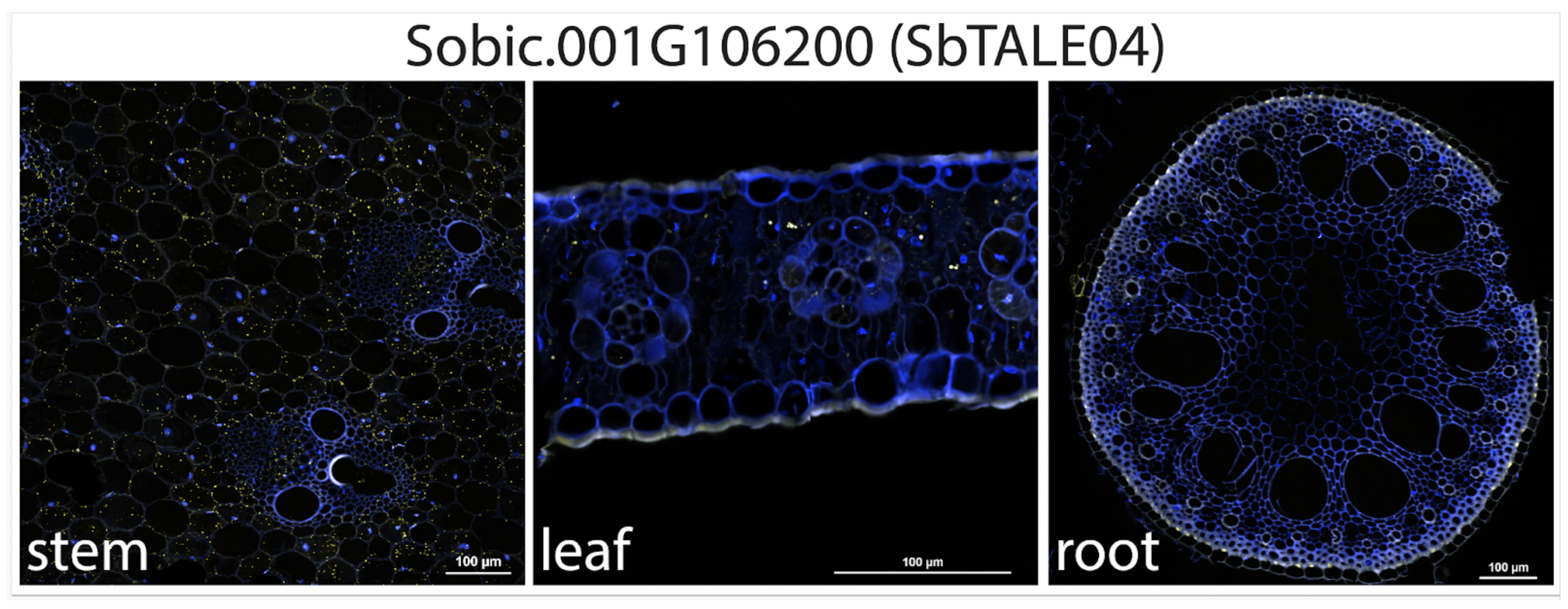
A genome-wide analysis of sorghum revealed that stems possess relatively few organ-specific genes due to their meristematic origins, with two KNOX-like transcription factors, SbTALE03 and SbTALE04, emerging as key stem-preferred regulators and promising tools for targeted engineering supported by regulatory and network evidence.
Keywords: Gene regulatory network, Organ specific, Sorghum, Stem, Stem specific, Transcription factor binding site
This study advances our understanding of stem-preferred gene regulation in sorghum and lays the foundation for developing targeted regulatory tools. It grew from a close collaboration among the Marshall-Colón, Mullet, Moose and Swaminathan labs—brought together through the vision and mentorship of the late Amy Marshall-Colón.
Sorghum bicolor is an emerging bioenergy crop valued for its deep root system, resilience to heat and drought, and high lignocellulosic and sugar yields stored in its tall stems. Although genomic resources for sorghum have expanded, comprehensive analyses of organ-specific transcriptional programs remain limited, particularly for the stem. Scientists from the University of Illinois Urbana-Champaign, the DOE Center for Advanced Bioenergy and Bioproducts Innovation and collaborating institutions conducted a genome-wide analysis of transcriptomes across major organs, which revealed that the stem contains far fewer organ-specific genes than leaves or roots, a pattern consistent with earlier reports. This disparity likely reflects developmental origins: leaves and roots arise from meristematic layers with highly specialized functions, whereas stems originate largely from the L3 lineage of the shoot apical meristem, whose derivatives contribute broadly to multiple shoot organs. Consequently, L3-derived transcriptional programs are more widely shared and less likely to yield strongly stem-specific signatures. Among the few truly stem-preferred genes identified, SbTALE03 and SbTALE04—homologs of maize KNOX genes—emerged as promising candidates. Their expression patterns were consistently stem-specific across diverse sorghum genotypes, suggesting potential utility as endogenous promoters for targeted transgene expression that avoids the drawbacks of constitutive promoters.
Regulatory analyses further revealed an enrichment of organ-specific cis-regulatory motifs within 300 bp upstream of the start codon, highlighting the functional importance of the core promoter and adjacent 5′ UTR in directing spatial gene expression. Gene regulatory network inference indicated that SbTALE03 and SbTALE04 function in a developmental-stage-dependent manner, shifting from roles in meristem maintenance during early growth to more specialized regulatory functions after floral initiation—patterns consistent with conserved KNOX family activities across species. Their evolutionary conservation as tandem duplicates in both sorghum and maize supports long-term subfunctionalization. Despite these insights, limitations remain due to the absence of sorghum-specific chromatin and TF-binding datasets, underscoring the need for future regulatory profiling to refine network predictions and deepen understanding of stem-specific gene control.
SorghumBase Examples:
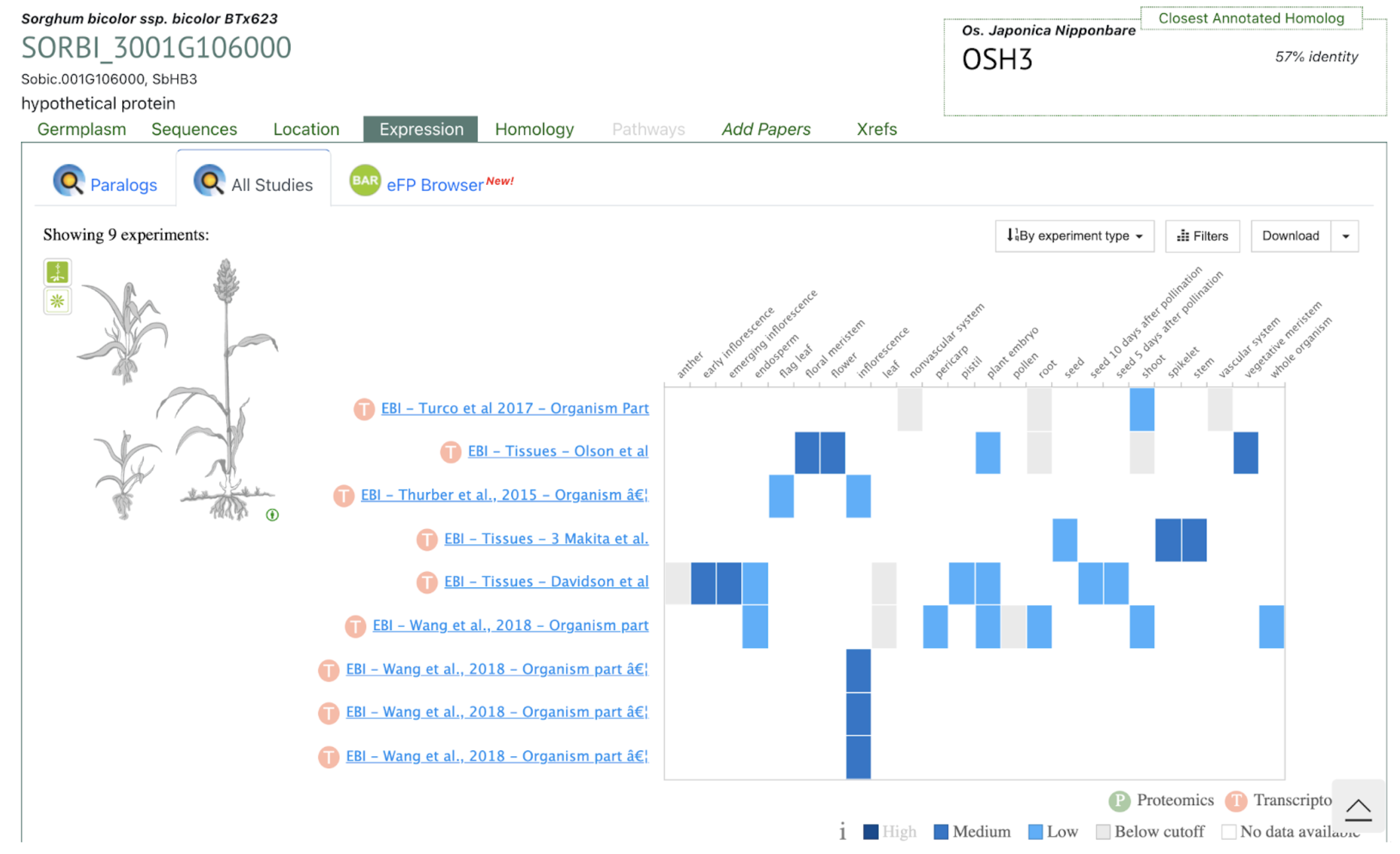
The SorghumBase Expression tab displays transcript abundance for SbTALE03 across nine datasets representing multiple tissues and developmental stages in BTx623. SbTALE03 shows distinctively elevated expression in stem-associated tissues, including developing internodes, peduncle, and early stem regions, while expression in root, leaf, and reproductive tissues remains low or below detection.
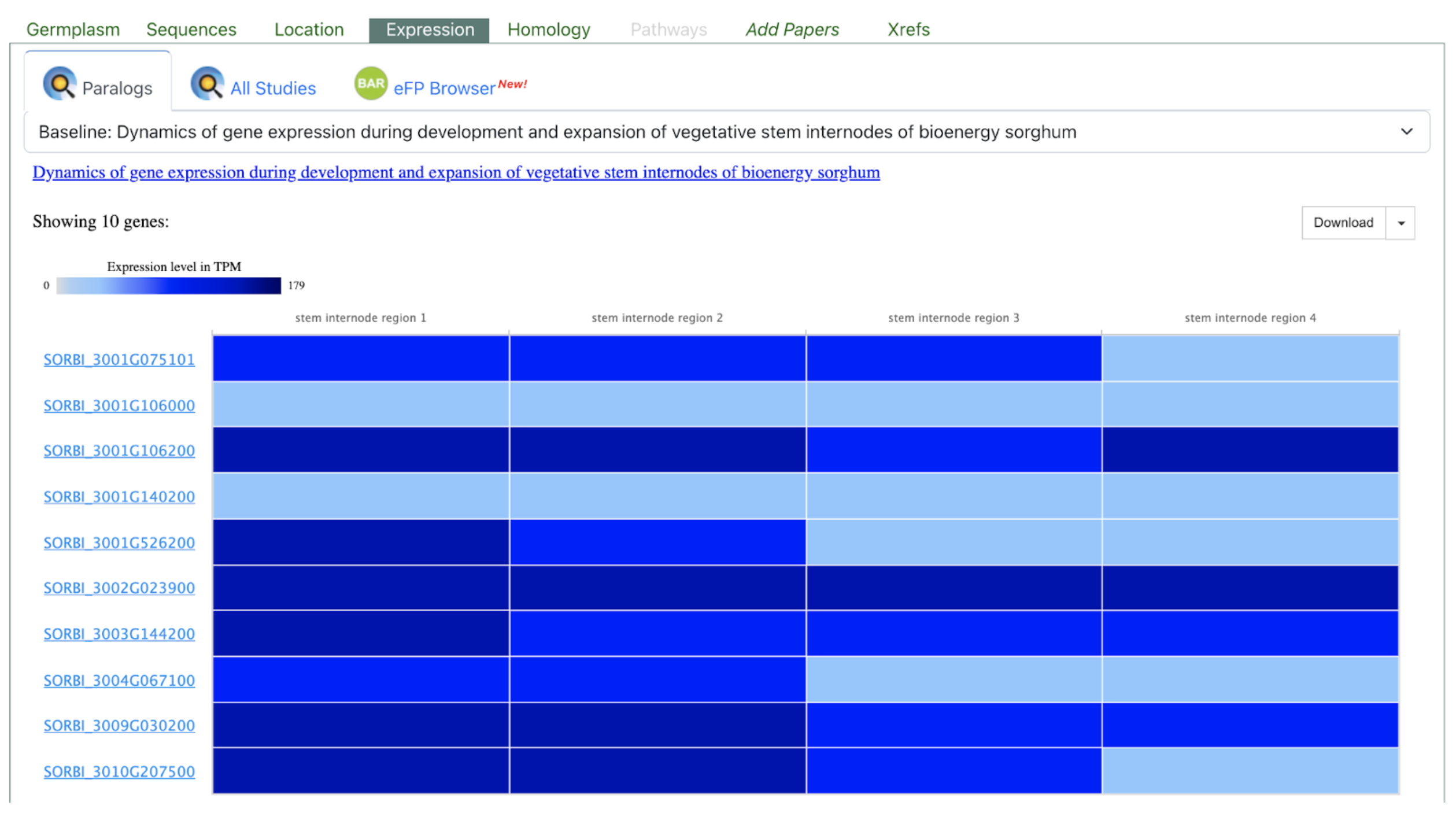
The SorghumBase Expression Atlas shows both genes within a paralog cluster associated with sorghum stem development. Although SbTALE03 and SbTALE04 share a common TALE-domain ancestry, their expression patterns differ markedly across the four stem internode regions shown. SbTALE03 displays moderate but consistent expression throughout the internode gradient, whereas SbTALE04 exhibits stronger and more dynamic expression, particularly in the more mature internode regions. This divergence suggests subfunctionalization of the two paralogs, consistent with the study’s findings that each gene regulates distinct components of the stem developmental gene regulatory network. These contrasting expression profiles highlight how SorghumBase can reveal functional differentiation among paralogs in sorghum developmental biology
Reference:
Fu J, James B, Hetti-Arachchilage M, Lei Y, McKinley B, Kurtz E, Barry K, Moose SP, Mullet JE, Swaminathan K, Marshall-Colon A. Stage-resolved gene regulatory network analysis reveals developmental reprogramming and genes with robust stem-preferred expression in sorghum. BMC Plant Biol. 2025 Oct 1;25(1):1275. PMID: 41034720. doi: 10.1186/s12870-025-07303-1. Read more