Conserved gene regulatory networks underpin nitrogen metabolism across plants, with dynamic and species-specific rewiring in maize and sorghum during rapid nitrogen deprivation and recovery, revealing key transcriptional regulators that shape nitrogen use efficiency and sustainable crop productivity.
Keywords: differentially expressed genes, feed‐forward loops, gene regulatory network, maize, nitrate, nitrogen uptake efficiency, sorghum, transcription factors
Deciphering the regulatory logic underlying rapid nitrogen responses in cereals, this work establishes a network-based framework for improving nitrogen use efficiency, with direct implications for enhancing crop performance while mitigating the environmental footprint of modern agriculture. – Braynen
Efficient nitrogen (N) use is central to sustainable agriculture, balancing the need for high crop productivity with the imperative to reduce environmental harm. Excessive N fertilizer application, while increasing yields, contributes to soil and water pollution, biodiversity loss, greenhouse gas emissions, and a substantial carbon footprint associated with industrial ammonia synthesis. Improving nitrogen use efficiency (NUE) therefore requires a detailed understanding of how crops acquire, assimilate, and respond to N availability at the molecular level. While most studies have focused on long-term N deprivation in model species such as Arabidopsis, maize, and rice, comparatively little is known about the early transcriptional responses to rapid changes in N status. Addressing this gap, the present work integrates gene regulatory network (GRN) analyses with short-term N deprivation and recovery experiments to dissect conserved and species-specific regulatory mechanisms controlling N metabolism in cereals.
Using an enhanced yeast one-hybrid (eY1H) networks derived from Arabidopsis and maize and projected onto sorghum, combined with spatiotemporal transcriptomics and Gini-correlation analyses, scientists from the USDA-ARS, Cold Spring Harbor Laboratory, National Research Council Canada, University of California Berkeley, University of California Davis, Corteva Agriscience and University of Saskatchewan identified conserved protein–DNA interactions and key transcription factor (TF) hubs involved in N signaling. The results reveal rapid and dynamic transcriptional reprogramming in maize and sorghum within hours of N deprivation, with notable tissue- and species-specific differences in N transport and carbon metabolism. Central regulators, including NLP17 and members of the bZIP family, emerge as highly connected hubs coordinating N uptake, assimilation, and signaling, often through complex feed-forward loop architectures. Despite a conserved core regulatory framework across dicots and monocots, fine-scale rewiring and temporal dynamics distinguish maize and sorghum strategies for restoring N homeostasis. Together, these findings advance our understanding of the conserved and adaptive regulatory logic underlying NUE and provide a framework for targeting regulatory networks to improve crop performance and environmental sustainability.
SorghumBase examples:
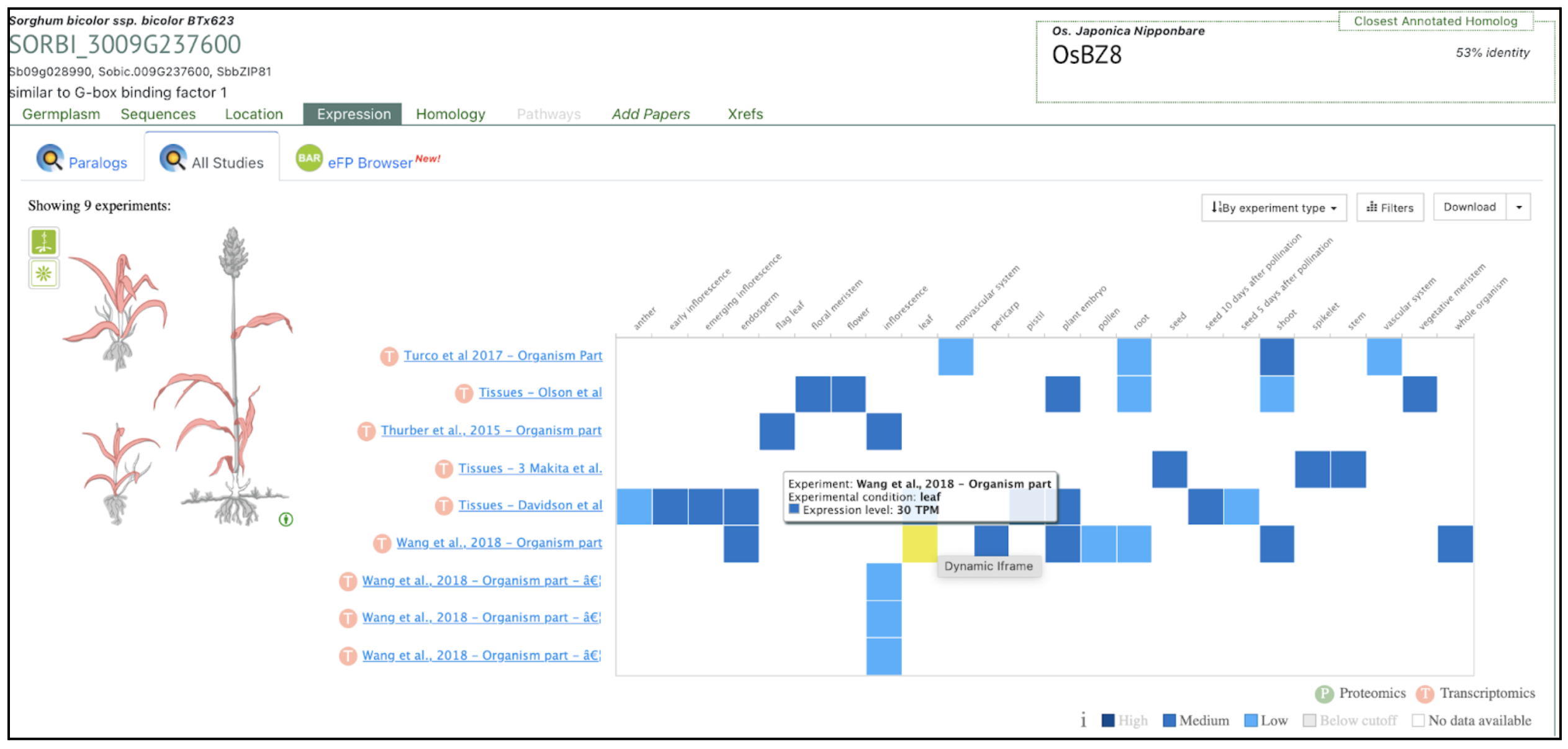
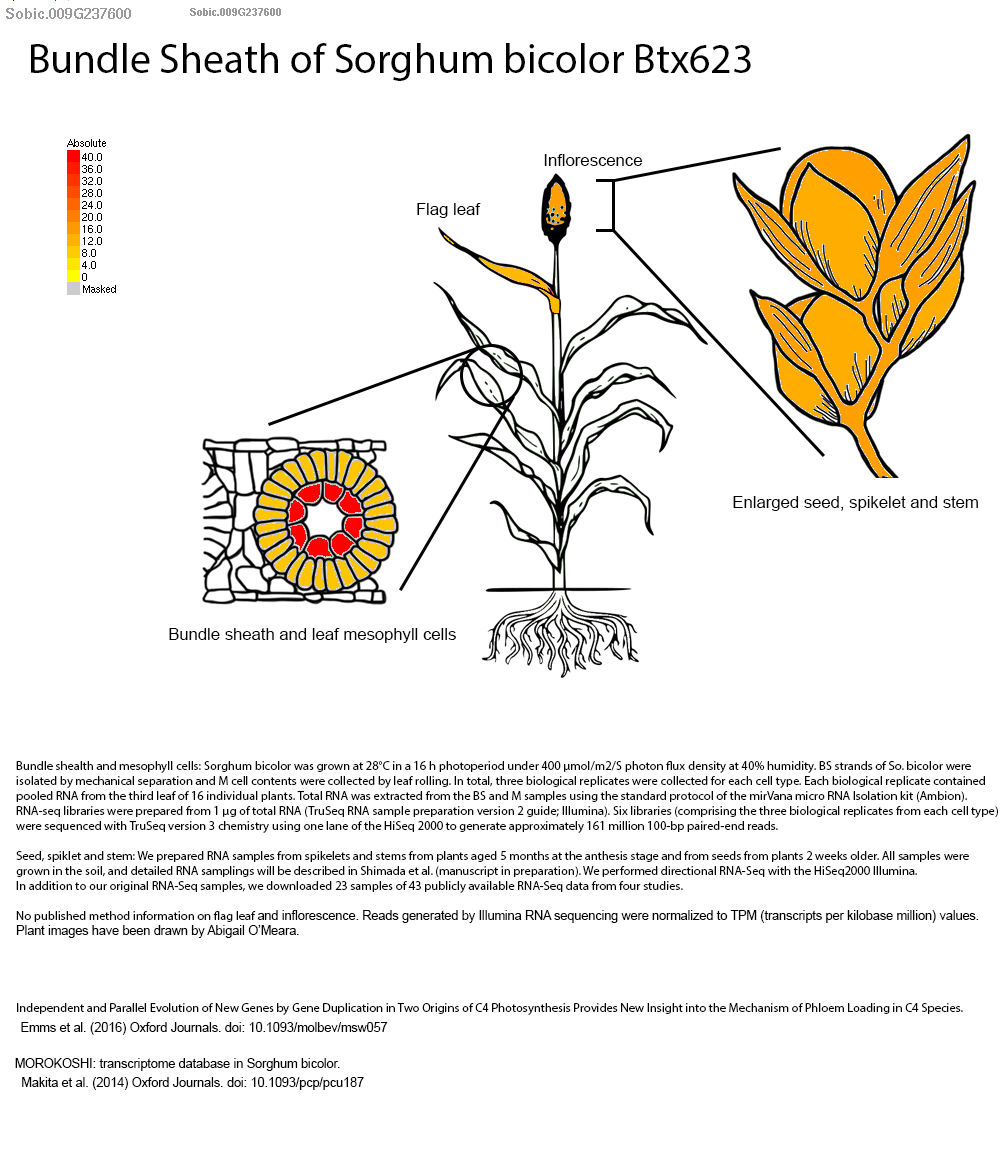
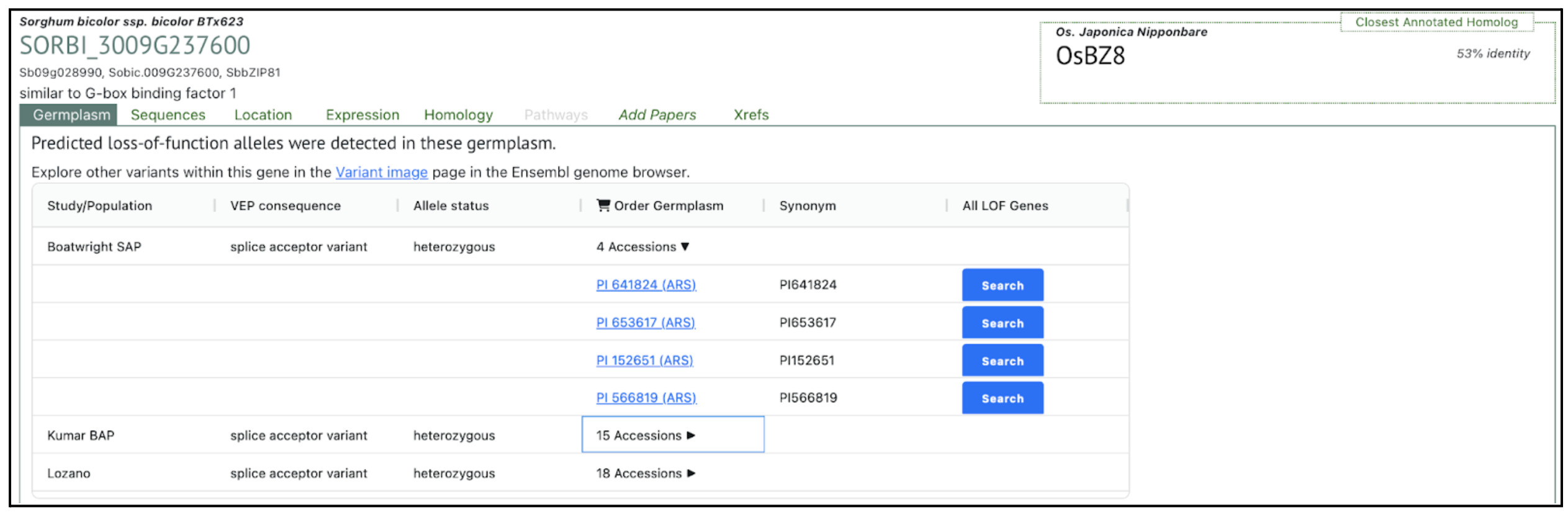
Reference:
Braynen J, Zhang L, Kumari S, Olson A, Kumar V, Regulski M, Liseron-Monfils C, Gaudinier A, Bågman AM, Abbitt S, Frank MJ, Shen B, Kochian L, Brady SM, Ware D. Decoding nitrogen uptake efficiency in maize and sorghum: insights from comparative gene regulatory networks. Plant J. 2025 Dec;124(6):e70631. PMID: 41411658. doi: 10.1111/tpj.70631. Read more
Related Project Websites:
- Ware Lab at Cold Spring Harbor Laboratory: https://warelab.cshl.edu/
