Drought tolerance in sorghum arises from coordinated molecular, biochemical, and physiological mechanisms, including elevated osmoprotectant levels, enhanced antioxidant defenses, and activation of ABA-dependent bZIP transcription factors that collectively maintain cellular stability and promote resilience under water stress.
Keywords: Antioxidant, Compatible osmolytes, Molecular responses, Physiological responses
Drought stress triggers complex molecular, physiological, and biochemical responses in plants, primarily regulated through transcriptional modulation of stress-responsive genes. Researchers from University of Guilan, Khuzestan Agricultural and Natural Resources, Research and Education Center and Charles University integrated transcriptomic and physiological analyses to identify key molecular mechanisms underlying drought resilience in sorghum. RNA-seq meta-analysis and gene expression validation in tolerant and sensitive sorghum cultivars revealed that drought stress substantially increased the accumulation of osmoprotectants such as proline and soluble sugars, which play critical roles in osmotic regulation and water retention. These changes were accompanied by elevated antioxidant enzyme activities, including catalase, peroxidase, superoxide dismutase, and ascorbate peroxidase, which collectively mitigated oxidative damage by scavenging reactive oxygen species. The tolerant cultivar displayed a more coordinated activation of these responses, suggesting a stronger capacity for maintaining cellular homeostasis under stress.
At the molecular level, drought stress upregulated several bZIP family transcription factors (e.g., SORBI_3003G332200, SORBI_3003G368300, and SORBI_3010G081800), known to mediate abscisic acid (ABA)-dependent regulatory pathways that enhance drought tolerance through transcriptional reprogramming of downstream stress-responsive genes. These findings highlight the synergistic role of osmotic adjustment, antioxidant defense, and transcriptional regulation in conferring drought resilience in sorghum. Although conducted under controlled greenhouse conditions, the study provides critical insights into the physiological, biochemical, and genetic foundations of drought tolerance. Future field-based and multi-omics investigations are recommended to validate these candidate genes, expand understanding of regulatory networks, and support the molecular breeding of sorghum cultivars with enhanced drought resilience.
SorghumBase Examples:
Authors have identified several bZIP family transcription factors (e.g., SORBI_3003G332200, SORBI_3003G368300, and SORBI_3010G081800), known to mediate abscisic acid (ABA)-dependent regulatory pathways that enhance drought tolerance through transcriptional reprogramming of downstream stress-responsive genes. We picked up this gene SORBI_3003G332200 to explore more on sorghumbase (https://sorghumbase.org/).
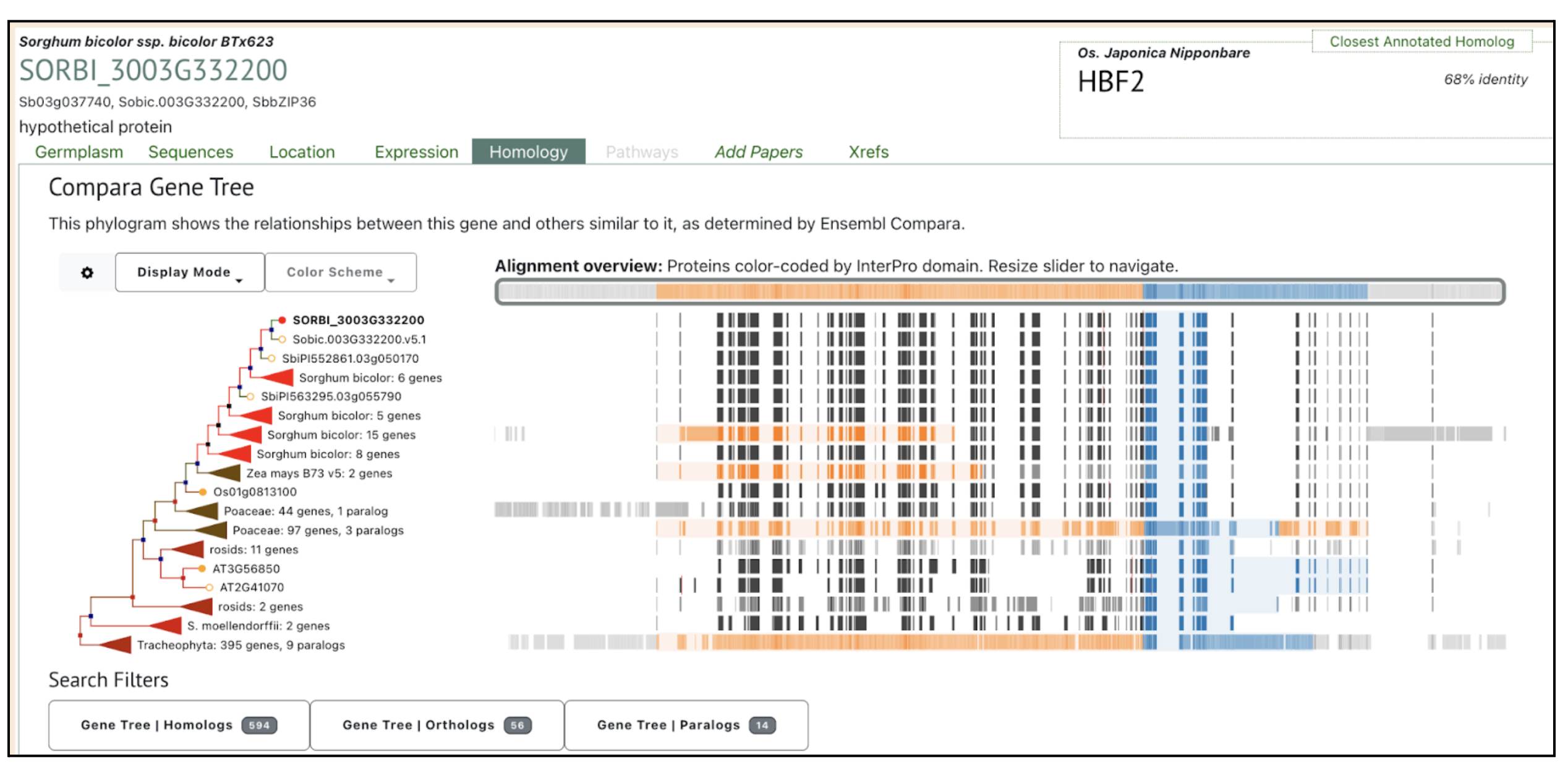
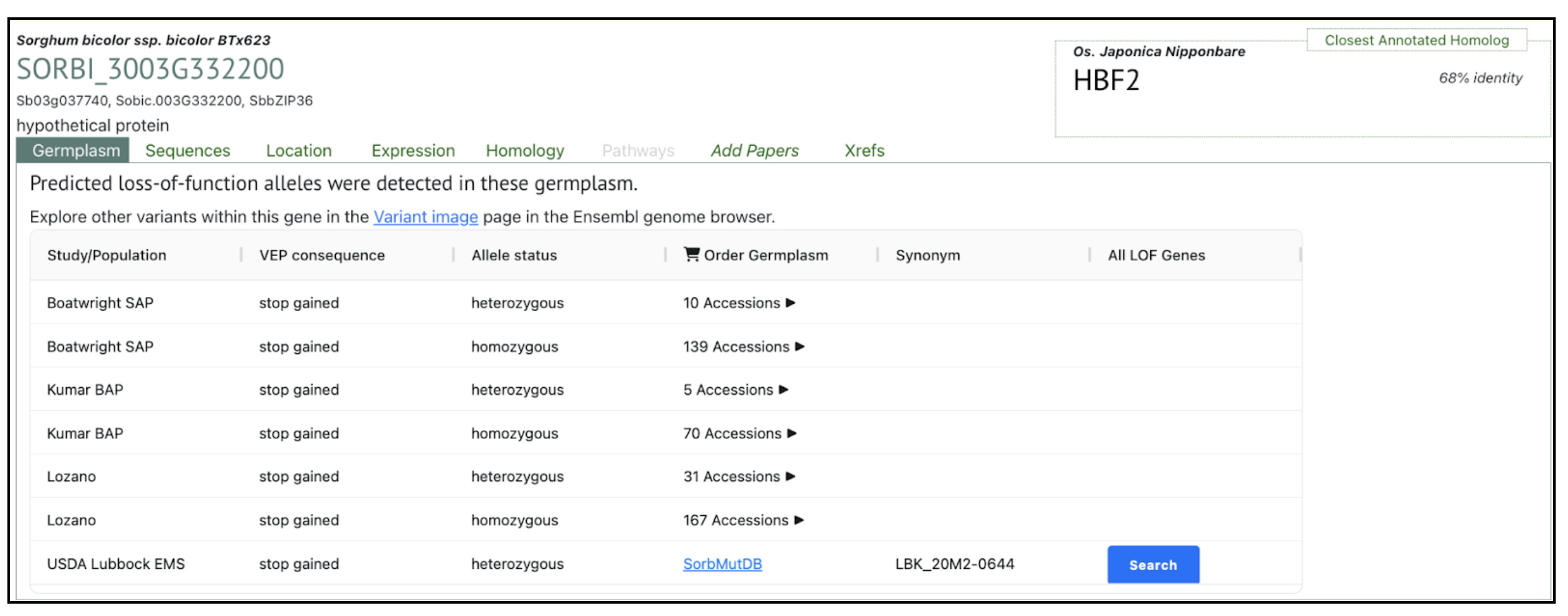
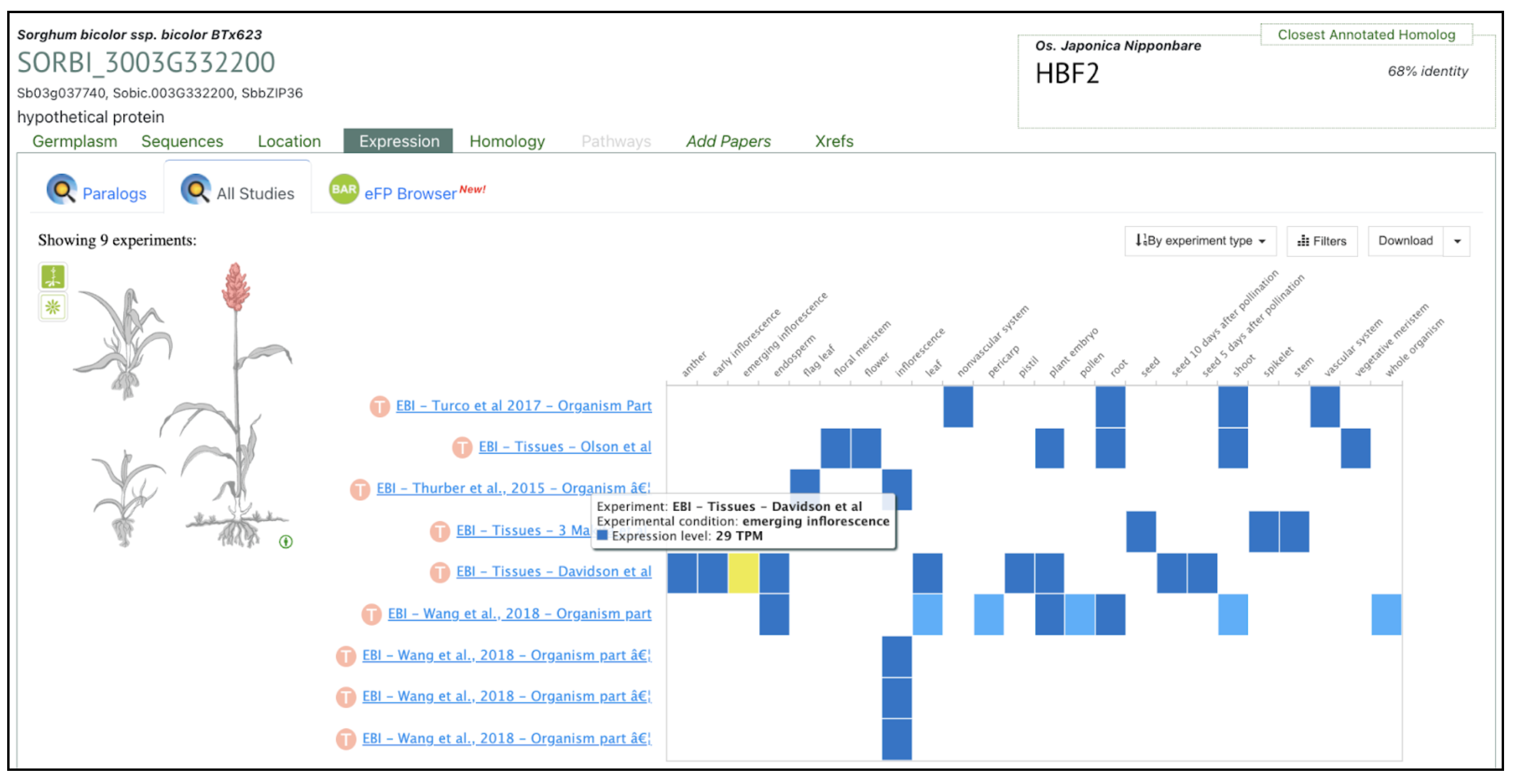
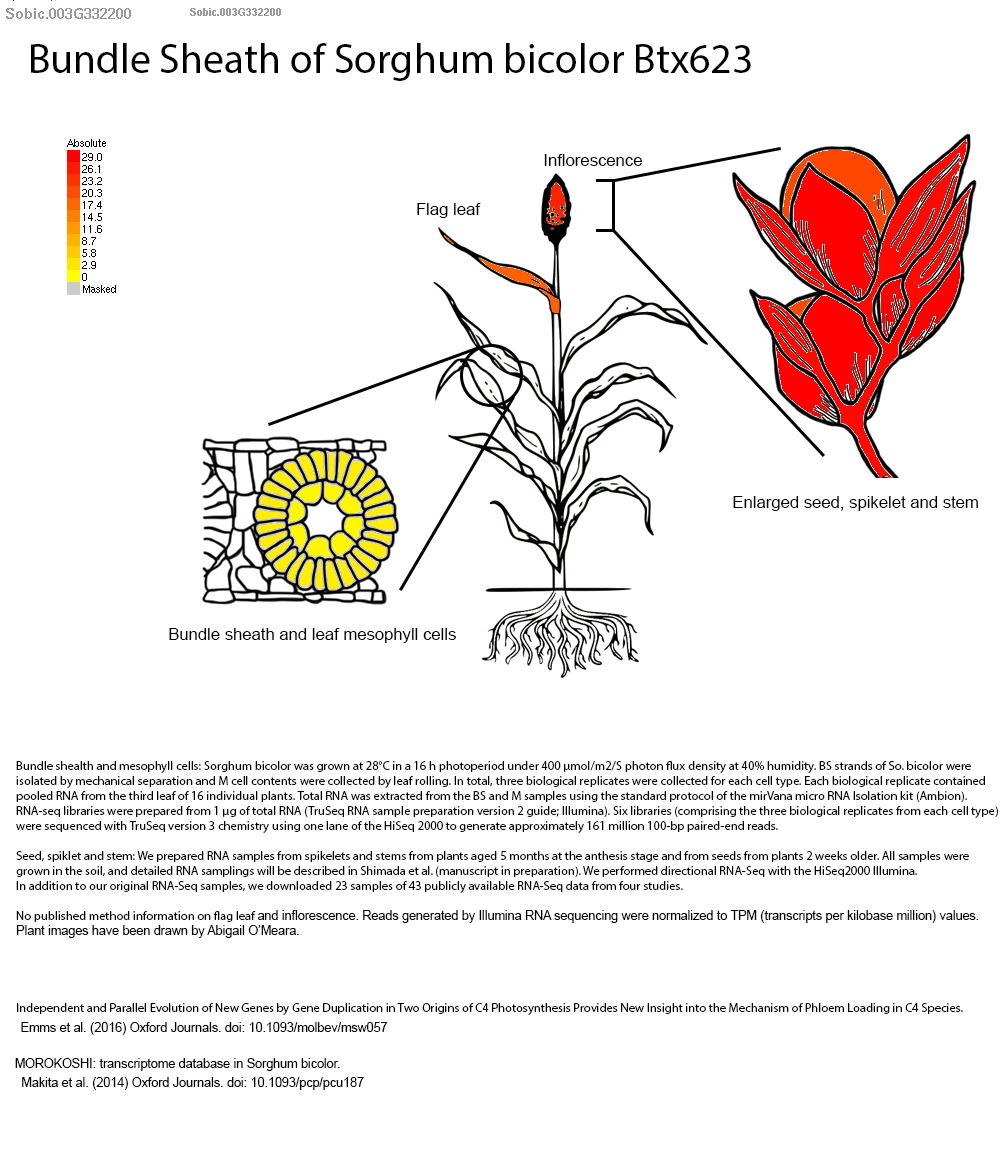
Kazemi H, Sabouri A, Aalami A, Abedi A, Nezamivand-Chegini M. A validation study by integrated analysis of physiological, biochemical, and meta-gene expression responses to drought stress in sorghum (Sorghum bicolor L.). Protoplasma. 2025 Sep 17. PMID: 40962917. doi: 10.1007/s00709-025-02112-7. Read more