Sorghum and maize descend from a common Poaceae ancestor and diverged from each other around 12 million years ago. The genes stemming from their independent domestication events show little overlap. During domestication, deleterious alleles often hitchhike along with the genes coding desirable traits and become fixed in the population. Understanding deleterious mutations in both species is important for optimizing these crops and is of great interest to scientists and breeders alike.
To enable investigation of differences between the domestication-related genes of sorghum and maize Lozano and colleagues at Cornell University in Ithaca, New York and the University of California Davis performed whole-genome resequencing to analyze approximately 13 million variants from 499 sorghum lines and compared the genetic variants with 25 million variants previously identified among 1,218 maize lines. The sorghum lines consist of the following population panels: 254 TERRA-MEPP accessions, 203 TERRA-REF accessions, and 42 accessions previously characterized by Mace et al (2013). Through comparing the deleterious mutations between the two species, the researchers showed that while maize analysis results were in line with the domestication-cost hypothesis, sorghum’s were not. Deleterious mutations in sorghum were not in significantly greater abundance in domesticated varieties when compared with wild types.
In addition, a gene deleterious index was constructed using the sorghum and maize data with an accuracy of greater than 0.5. Through comparing the evolutionary genetics of sorghum and maize, the researchers provide a deeper understanding of the evolutionary dynamics of deleterious mutations. Their comparative research helps in the identification of deleterious mutations that can ultimately be targeted for removal.
“Gaining a deeper understanding of the evolutionary patterns of deleterious mutations in a comparative genomics framework may help us to more effectively breed both sorghum and maize” – Gore
SorghumBase examples:
These data can be accessed at SorghumBase in several ways. For example, below is a table of variants and gene neighborhood view in Sorghumbase for SORBI_3006G108000, the sorghum ortholog of rice dw2. The table provides information on the variants within the gene, including the alleles, source study and consequence type, while the gene neighborhood view provides insights into local gene gain and loss.
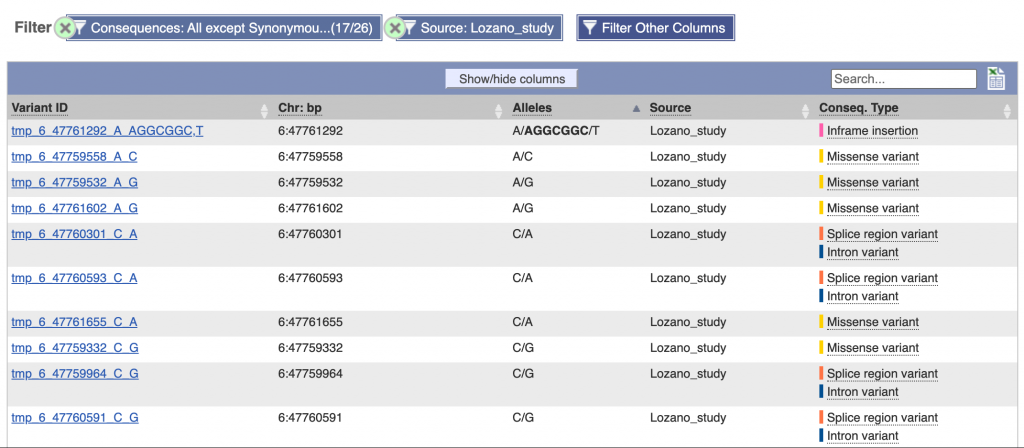
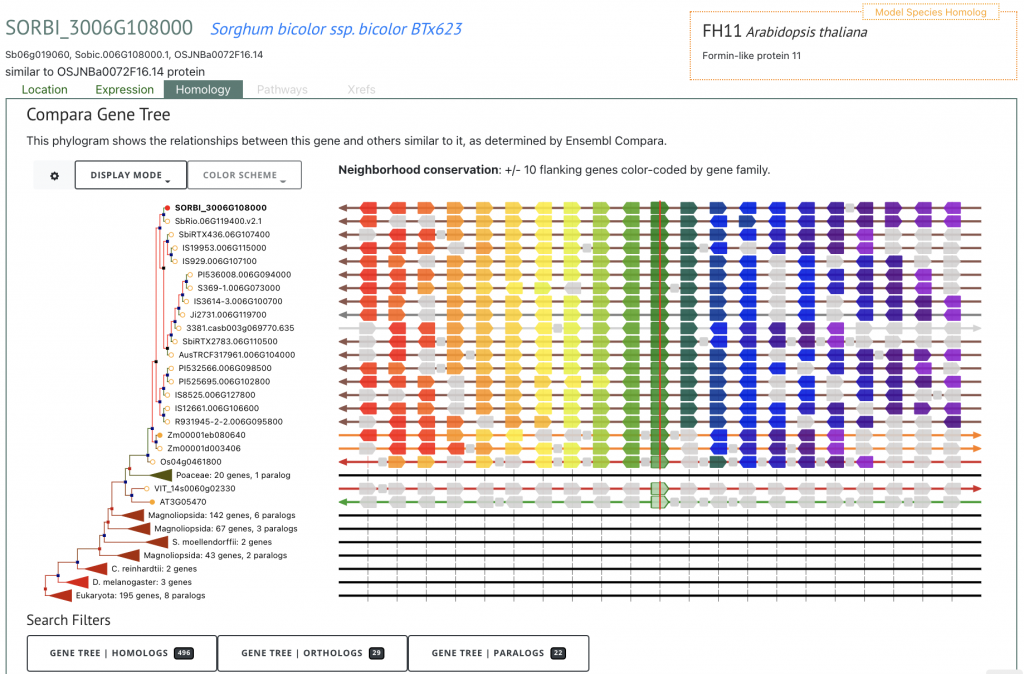
Reference
Lozano, R., Gazave, E., dos Santos, J.P.R., Stetter, M.G., Valluru, R., Bandillo, N., Fernandes, S.B., Brown, P.J., Shakoor, N., Mockler, T.C., Cooper, E.A., Perkins, M.T., Buckler, E.S., Ross-Ibarra, J., and Gore, M.A. Comparative evolutionary genetics of deleterious load in sorghum and maize. Nat. Plants 7, 17–24 (2021). PMID: 33452486. DOI: 10.1038/s41477-020-00834-5. Read more
Mace ES, Tai S, Gilding EK, Li Y, Prentis PJ, Bian L, Campbell BC, Hu W, Innes DJ, Han X, Cruickshank A, Dai C, Frère C, Zhang H, Hunt CH, Wang X, Shatte T, Wang M, Su Z, Li J, Lin X, Godwin ID, Jordan DR, Wang J. Whole-genome sequencing reveals untapped genetic potential in Africa’s indigenous cereal crop sorghum. Nat Commun. 2013;4:2320. PMID: 23982223. DOI: 10.1038/ncomms3320.
Related Project Websites
- Cornell University The Gore Lab https://blogs.cornell.edu/gorelab/
- University of Arizona TERRA-REF https://terraref.org/
- University of Illinois TERRA-MEPP https://terra-mepp.illinois.edu/
