Leaf angle is a trait that affects how closely packed plants can be grown, the amount of sunlight that falls on the leaves, photosynthetic rate, and yield. Sorghum plants tend to have more horizontal leaves (with larger angles) towards the top of the plant and vertical leaves (with smaller angles) towards the bottom. This is in stark contrast to the “Smart Canopy” Theory, which states that vertical leaves should be towards the top of the plant and horizontal leaves towards the bottom, allowing for more light to reach the bottom leaves and more efficient light interception and conversion for the entire plant. The potential of altering the leaf angle to significantly increase yield is of great interest to scientists and growers.
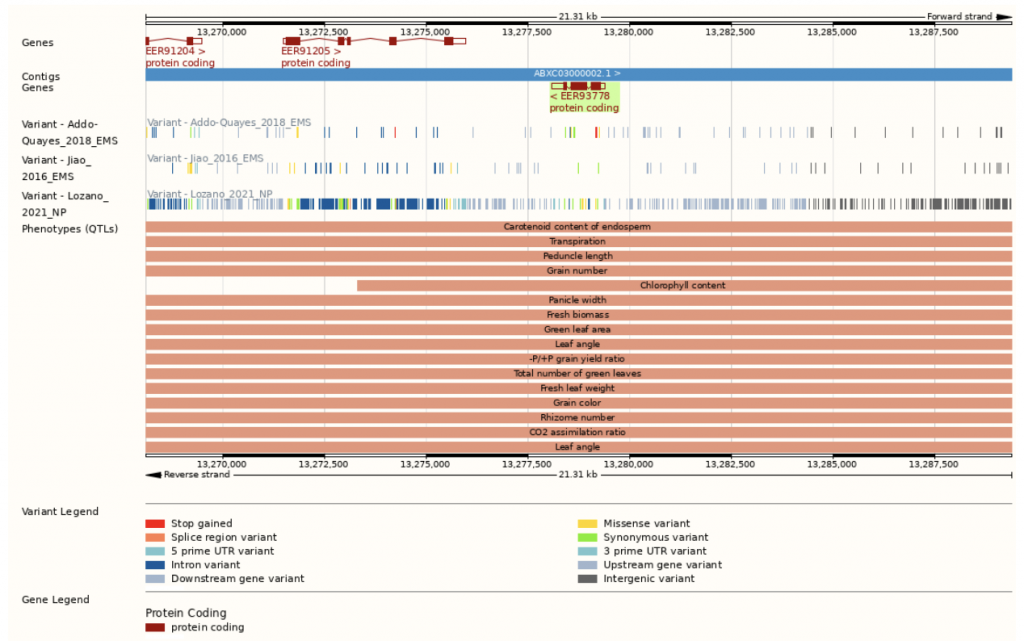
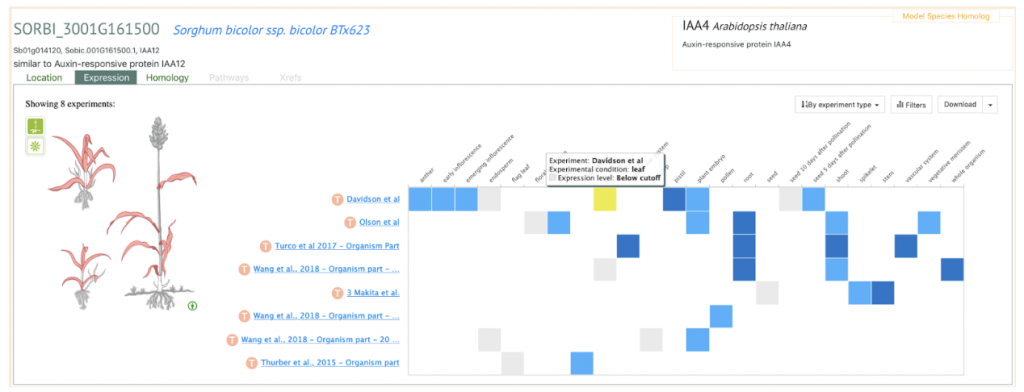
In an effort to determine the genetic factors behind leaf angle of whole sorghum plants, Natukunda and colleagues from Iowa State University and the USDA performed RNA seq on collar tissue from different canopy levels. Cells in this collar tissue determine leaf angle. The list of differentially expressed genes (DEGs) was narrowed to 284 genes based on co-localization with 69 known leaf angle QTLs. A list of 11 candidate genes was derived from these by considering consistency of gene expression in different genotypes, canopy layer phenotype patterns, homology, and putative function. Seven of these genes were known to influence leaf angle in rice and maize and the remaining four are involved in processes that ultimately affect leaf angle, such as cell size, shape or number. These candidates should be researched for the effect they have on tissues at various points in sorghum development, as they might have pleiotropic effects, before they are used in genetic engineering in an attempt to alter the leaf angle phenotype.
“In sorghum, leaf angle distribution across the canopy is opposite to the photosynthetically efficient “Smart Canopy” ideotype. Therefore, this study was designed to discover genes differentially expressed across canopy layers in collar tissues, whose cells determine leaf angle. Multiple sources of evidence facilitated the identification of a subset of promising candidate genes that could be exploited to develop optimized canopies with higher photosynthetic capacity and thus yield. Current research in our lab is dedicated to the functional validation of these candidates and to determine their possible pleiotropic effects. A similar approach can be utilized to improve leaf angle distribution in other grasses.” M. Ibore-Natukunda and M.G. Salas-Fernandez
SorghumBase examples:
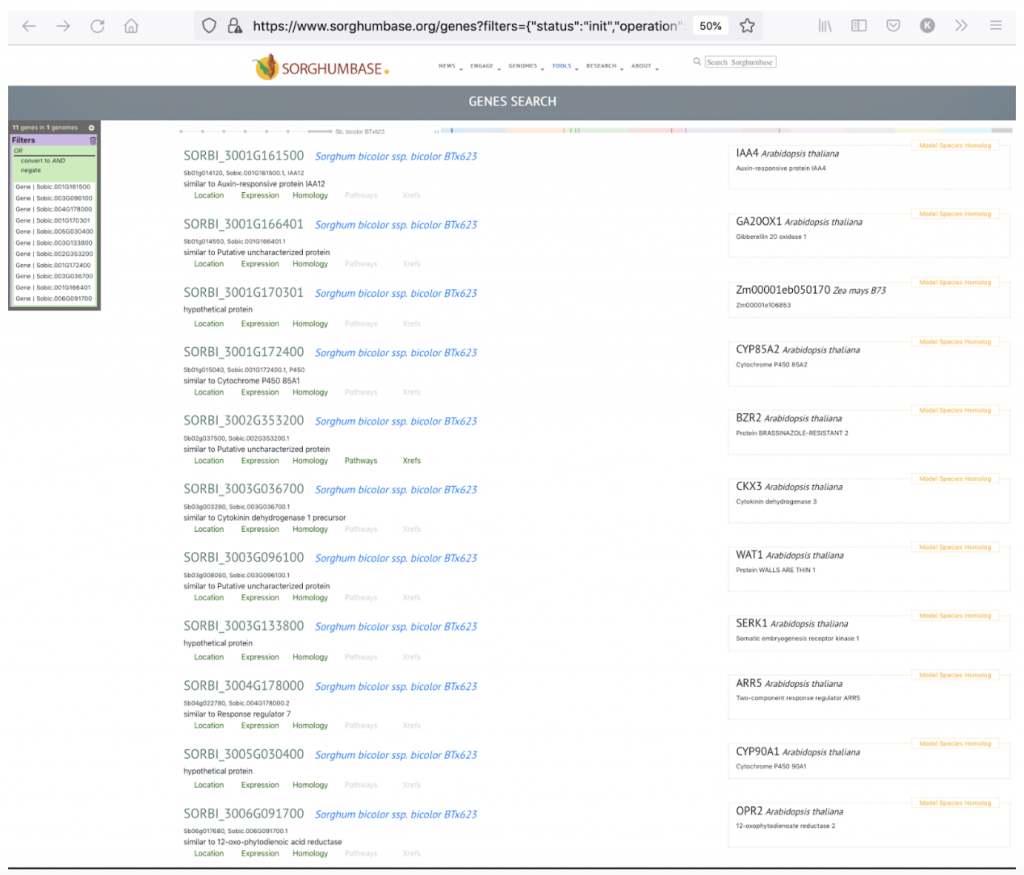
The 11 candidate genes for leaf angle may be seen in the following link:
- Sobic.001G161500
- Sobic.003G096100
- Sobic.004G178000
- Sobic.001G170301
- Sobic.005G030400
- Sobic.003G133800
- Sobic.002G353200
- Sobic.001G172400
- Sobic.003G036700
- Sobic.001G166401
- Sobic.006G091700

References
Davidson RM, Gowda M, Moghe G, Lin H, Vaillancourt B, Shiu SH, Jiang N, Robin Buell C. Comparative transcriptomics of three Poaceae species reveals patterns of gene expression evolution. Plant J. 2012 Aug;71(3):492-502. PMID: 22443345. DOI: 10.1111/j.1365-313X.2012.05005.x.
Natukunda MI, Mantilla-Perez MB, Graham MA, Liu P, Salas-Fernandez MG. Dissection of canopy layer-specific genetic control of leaf angle in Sorghum bicolor by RNA sequencing. BMC Genomics. 2022 Feb 3;23(1):95. DOI: 10.1186/s12864-021-08251-4. PMID: 35114939. Read More
Wang B, Regulski M, Tseng E, Olson A, Goodwin S, McCombie WR, Ware D. A comparative transcriptional landscape of maize and sorghum obtained by single-molecule sequencing. Genome Res. 2018 Jun;28(6):921-932. PMID: 29712755. DOI: 10.1101/gr.227462.117.
Related Project Websites
Salas-Fernandez Lab:
https://www.agron.iastate.edu/people/maria-salas-fernandez
https://www.plantbreeding.iastate.edu/projects
Twitter: @ISU_Sorghum
M. Ibore’s twitter: @IboreMartha




