Genetic variation in crops is critical for robustness of the species and can be leveraged for creating improved cultivars. Association mapping population panels help identify the connections between genetic variations and phenotypic differences by comparing genomes from varied plant accessions that have evolved in a diverse array of environments. The sorghum association panel (SAP) includes lines from temperate climates, as well as photoperiod-insensitive lines from tropical climates. Although previously only portions of the genomes in the SAP were sequenced, reductions in the cost of this technology have allowed whole-genome sequencing (WGS). This has increased the identification of genetic diversity and can be used with genome-wide association mapping (GWAS) for genomic selection with greater specificity. The identification of additional causal variants in sorghum, which can be used to improve lines, is of great interest to researchers, breeders, and growers.
Boatwright and colleagues report the generation of a resource consisting of approximately 44 million genetic variants of diverse types called using whole-genome sequencing (WGS) of the 400 SAP accessions in a June 2022 publication.
The variants include approximately 38 million single-nucleotide polymorphisms (SNPs), about 5 million indels, and ~170,000 copy-number variations (CNVs). These markers represent a major increase in the density and variant types over previous resources.
To demonstrate the superior value of this WGS-based resource relative to existing GBS-based resources, the authors identified several putatively novel genomic associations for plant height and tannin content in addition to confirming well known height loci Dw1 (SORBI_3009G229800), Dw2 (SORBI_3006G067700) and Dw3 (SORBI_307G163800), and tannin content locus Tan 1 (SORBI_3004G280800). Such novel associations spanning two loci for plant height and four loci for tannin content were not identified previously using lower-density GBS marker sets. Furthermore, the authors identified 18 selection peaks across subpopulations that formed due to evolutionary divergence during domestication, and found six Fst peaks resulting from comparisons between converted tropical lines and temperately adapted breeding lines within the SAP that were distinct from the peaks associated with historic selection.
The SAP population has been and continues to serve as a significant public resource for sorghum research. SorghumBase will be working with the authors to integrate access to the SAP variation in the context of Sorghum BTx623 reference genome in 2022.
“By providing these high-throughput genomic resources based on the sorghum association panel, we hope to build upon the work established by the sorghum community in a way that expands the breadth and depth of future studies. We anticipate that these high-density variants of diverse types will enhance the dissection and improvement of the genetic architecture underlying vital agronomic, physiological, and compositional traits in sorghum.” -Lucas Boatwright
“Like many other investigators of the sorghum crop improvement community, we strive to generate community resources that will be of tangible value nationally and globally. This is another contribution to that coordinated effort.” -Stephen Kresovich
SorghumBase examples
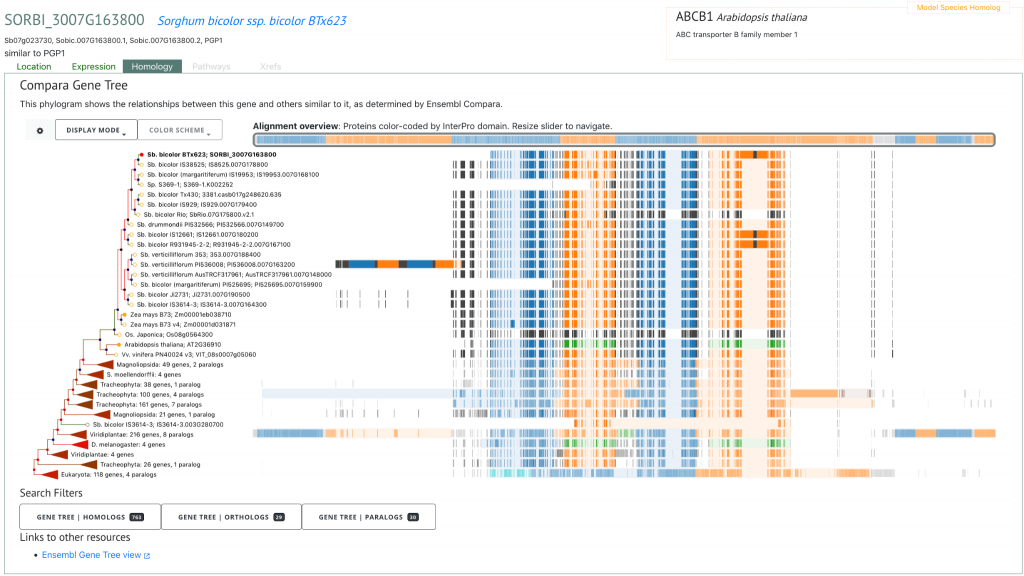
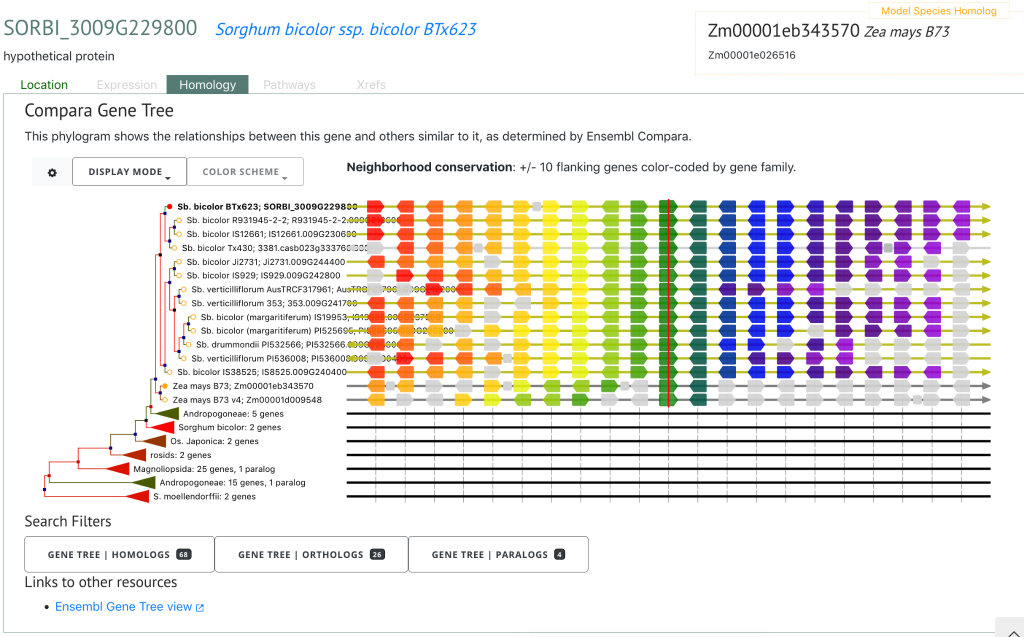
Reference
Boatwright JL, Sapkota S, Jin H, Schnable JC, Brenton Z, Boyles R, Kresovich S. Sorghum Association Panel whole-genome sequencing establishes cornerstone resource for dissecting genomic diversity. Plant J. 2022 Jun 2. PMID: 35653240. DOI: 10.1111/tpj.15853. Read more
Related Project Websites:
- https://linktr.ee/LucasBoatwright
- https://www.skgenomics.com/
- https://ilci.cornell.edu/our-team/stephen-kresovich/

